Stability and kinetics of G-quadruplex structures
- PMID: 18718931
- PMCID: PMC2553573
- DOI: 10.1093/nar/gkn517
Stability and kinetics of G-quadruplex structures
Abstract
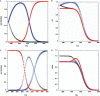
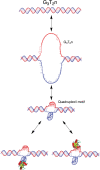
In this review, we give an overview of recent literature on the structure and stability of unimolecular G-rich quadruplex structures that are relevant to drug design and for in vivo function. The unifying theme in this review is energetics. The thermodynamic stability of quadruplexes has not been studied in the same detail as DNA and RNA duplexes, and there are important differences in the balance of forces between these classes of folded oligonucleotides. We provide an overview of the principles of stability and where available the experimental data that report on these principles. Significant gaps in the literature have been identified, that should be filled by a systematic study of well-defined quadruplexes not only to provide the basic understanding of stability both for design purposes, but also as it relates to in vivo occurrence of quadruplexes. Techniques that are commonly applied to the determination of the structure, stability and folding are discussed in terms of information content and limitations. Quadruplex structures fold and unfold comparatively slowly, and DNA unwinding events associated with transcription and replication may be operating far from equilibrium. The kinetics of formation and resolution of quadruplexes, and methodologies are discussed in the context of stability and their possible biological occurrence.
Figures
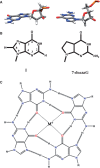

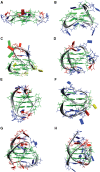

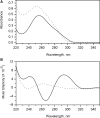
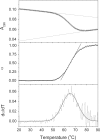

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

