Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice
- PMID: 18718932
- PMCID: PMC2566925
- DOI: 10.1093/pcp/pcn123
Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice
Abstract
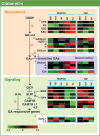
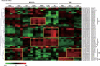
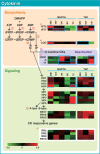
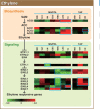
To investigate the involvement of phytohormones during rice microspore/pollen (MS/POL) development, endogenous levels of IAA, gibberellins (GAs), cytokinins (CKs) and abscisic acid (ABA) in the mature anther were analyzed. We also analyzed the global expression profiles of genes related to seven phytohormones, namely auxin, GAs, CKs, brassinosteroids, ethylene, ABA and jasmonic acids, in MS/POL and tapetum (TAP) using a 44K microarray combined with a laser microdissection technique (LM-array analysis). IAA and GA(4) accumulated in a much higher amount in the mature anther compared with the other tissues, while CKs and ABA did not. LM-array analysis revealed that sets of genes required for IAA and GA synthesis were coordinately expressed during the later stages of MS/POL development, suggesting that these genes are responsible for the massive accumulation of IAA and GA(4) in the mature anther. In contrast, genes for GA signaling were preferentially expressed during the early developmental stages of MS/POL and throughout TAP development, while their expression was down-regulated at the later stages of MS/POL development. In the case of auxin signaling genes, such mirror-imaged expression observed in GA synthesis and signaling genes was not observed. IAA receptor genes were mostly expressed during the late stages of MS/POL development, and various sets of AUX/IAA and ARF genes were expressed during the different stages of MS/POL or TAP development. Such cell type-specific expression profiles of phytohormone biosynthesis and signaling genes demonstrate the validity and importance of analyzing the expression of phytohormone-related genes in individual cell types independently of other cells/tissues.
Figures







References
-
- Ariizumi T, Kawanabe T, Hatakeyama K, Sato S, Kato T, Tabata S, et al. Ultrastructural characterization of exine development of the transient defective exine 1 mutant suggests the existence of a factor involved in constructing reticulate exine architecture from sporopollenin aggregates. Plant Cell Physiol. 2008;49:58–67. - PubMed
-
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

