Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes
- PMID: 18719128
- PMCID: PMC2527885
- DOI: 10.1073/pnas.0803626105
Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes
Abstract
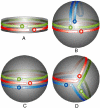
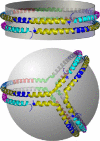
Spherical high density lipoproteins (HDL) predominate in human plasma. However, little information exists on the structure of the most common HDL protein, apolipoprotein (apo) A-I, in spheres vs. better studied discoidal forms. We produced spherical HDL by incubating reconstituted discoidal HDL with physiological plasma-remodeling enzymes and compared apoA-I structure in discs and spheres of comparable diameter (79-80 and 93-96 A). Using cross-linking chemistry and mass spectrometry, we determined that the general structural organization of apoA-I was overall similar between discs and spheres, regardless of diameter. This was the case despite the fact that the 93 A spheres contained three molecules of apoA-I per particle compared with only two in the discs. Thus, apoA-I adopts a consistent general structural framework in HDL particles-irrespective of shape, size and the number of apoA-Is present. Furthermore, a similar cross-linking pattern was demonstrated in HDL particles isolated from human serum. We propose the first experiment-based molecular model of apoA-I in spherical HDL particles. This model provides a new foundation for understanding how apoA-I structure modulates HDL function and metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Curtiss LK, Valenta DT, Hime NJ, Rye KA. What is so special about apolipoprotein AI in reverse cholesterol transport? Arterioscler Thromb Vasc Biol. 2006;26:12–19. - PubMed
-
- Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: A critical analysis and review. Biochim Biophys Acta. 2001;1531:4–46. - PubMed
-
- Segrest JP, et al. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. - PubMed
-
- Jonas A. In: Biochemistry of Lipids, Lipoproteins and Membranes. Vance DE, Vance JE, editors. Vol. 483. Elsevier Science; 2002. p. 504.
Publication types
MeSH terms
Substances
Grants and funding
- P60 DK020593/DK/NIDDK NIH HHS/United States
- DK58404/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 HL67093/HL/NHLBI NIH HHS/United States
- R01 HL48148/HL/NHLBI NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- K99/R00 HL1004925/HL/NHLBI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- DK20593/DK/NIDDK NIH HHS/United States
- R01 HL067093/HL/NHLBI NIH HHS/United States
- P20 RR016475/RR/NCRR NIH HHS/United States
- P20RR16475/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

