lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo
- PMID: 18719247
- PMCID: PMC2553750
- DOI: 10.1261/rna.1094208
lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo
Abstract
The poly(A) tail is a crucial determinant in the control of both mRNA translation and decay. Poly(A) tail length dictates the triggering of the degradation of the message body in the major 5' to 3' and 3' to 5' mRNA decay pathways of eukaryotes. In the 5' to 3' pathway oligoadenylated but not polyadenylated mRNAs are selectively decapped in vivo, allowing their subsequent degradation by 5' to 3' exonucleolysis. The conserved Lsm1p-7p-Pat1p complex is required for normal rates of decapping in vivo, and the purified complex exhibits strong binding preference for oligoadenylated RNAs over polyadenylated or unadenylated RNAs in vitro. In the present study, we show that two lsm1 mutants produce mutant complexes that fail to exhibit such higher affinity for oligoadenylated RNA in vitro. Interestingly, these mutant complexes are normal with regard to their integrity and retain the characteristic RNA binding properties of the wild-type complex, namely, binding near the 3'-end of the RNA, having higher affinity for unadenylated RNAs that carry U-tracts near the 3'-end over those that do not and exhibiting similar affinities for unadenylated and polyadenylated RNAs. Yet, these lsm1 mutants exhibit a strong mRNA decay defect in vivo. These results underscore the importance of Lsm1p-7p-Pat1p complex-mRNA interaction for mRNA decay in vivo and imply that the oligo(A) tail mediated enhancement of such interaction is crucial in that process.
Figures
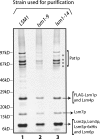
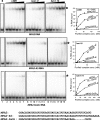
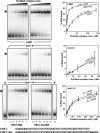

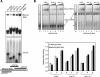

References
-
- Anderson, J.T. RNA turnover: Unexpected consequences of being tailed. Curr. Biol. 2005;15:R635–R638. - PubMed
-
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., Parker, R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
