Epigenetic reprogramming by adenovirus e1a
- PMID: 18719284
- PMCID: PMC2693122
- DOI: 10.1126/science.1155546
Epigenetic reprogramming by adenovirus e1a
Abstract
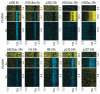
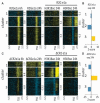
Adenovirus e1a induces quiescent human cells to replicate. We found that e1a causes global relocalization of the RB (retinoblastoma) proteins (RB, p130, and p107) and p300/CBP histone acetyltransferases on promoters, the effect of which is to restrict the acetylation of histone 3 lysine-18 (H3K18ac) to a limited set of genes, thereby stimulating cell cycling and inhibiting antiviral responses and cellular differentiation. Soon after expression, e1a binds transiently to promoters of cell cycle and growth genes, causing enrichment of p300/CBP, PCAF (p300/CBP-associated factor), and H3K18ac; depletion of RB proteins; and transcriptional activation. e1a also associates transiently with promoters of antiviral genes, causing enrichment for RB, p130, and H4K16ac; increased nucleosome density; and transcriptional repression. At later times, e1a and p107 bind mainly to promoters of development and differentiation genes, repressing transcription. The temporal order of e1a binding requires its interactions with p300/CBP and RB proteins. Our data uncover a defined epigenetic reprogramming leading to cellular transformation.
Figures
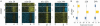
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

