Differential screening identifies transcripts with depot-dependent expression in white adipose tissues
- PMID: 18721461
- PMCID: PMC2547859
- DOI: 10.1186/1471-2164-9-397
Differential screening identifies transcripts with depot-dependent expression in white adipose tissues
Abstract
Background: The co-morbidities of obesity are tied to location of excess fat in the intra-abdominal as compared to subcutaneous white adipose tissue (WAT) depot. Genes distinctly expressed in WAT depots may impart depot-dependent physiological functions. To identify such genes, we prepared subtractive cDNA libraries from murine subcutaneous (SC) or intra-abdominal epididymal (EP) white adipocytes.
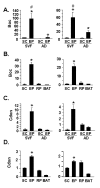
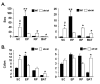
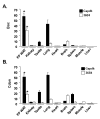
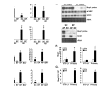
Results: Differential screening and qPCR validation identified 7 transcripts with 2.5-fold or greater enrichment in EP vs. SC adipocytes. Boc, a component of the hedgehog signaling pathway demonstrated highest enrichment (approximately 12-fold) in EP adipocytes. We also identified a dramatic enrichment in SC adipocytes vs. EP adipocytes and in SC WAT vs. EP WAT for transcript(s) for the major urinary proteins (Mups), small secreted proteins with pheromone functions that are members of the lipocalin family. Expression of Boc and Mup transcript was further assessed in murine tissues, adipogenesis models, and obesity. qPCR analysis reveals that EP WAT is a major site of expression of Boc transcript. Furthermore, Boc transcript expression decreased in obese EP WAT with a concomitant upregulation of Boc transcript in the obese SC WAT depot. Assessment of the Boc binding partner Cdon in adipose tissue and cell fractions thereof, revealed transcript expression similar to Boc; suggestive of a role for the Boc-Cdon axis in WAT depot function. Mup transcripts were predominantly expressed in liver and in the SC and RP WAT depots and increased several thousand-fold during differentiation of primary murine preadipocytes to adipocytes. Mup transcripts were also markedly reduced in SC WAT and liver of ob/ob genetically obese mice compared to wild type.
Conclusion: Further assessment of WAT depot-enriched transcripts may uncover distinctions in WAT depot gene expression that illuminate the physiological impact of regional adiposity.
Figures


References
-
- Coppack SW. Pro-inflammatory cytokines and adipose tissue. Proc Nutr Soc. 2001;60:349–356. - PubMed
-
- Gregoire FM. Adipocyte differentiation: from fibroblast to endocrine cell. Exp Biol Med (Maywood) 2001;226:997–1002. - PubMed
-
- Shimomura I, Funahashi T, Takahashi M, Maeda K, Kotani K, Nakamura T, Yamashita S, Miura M, Fukuda Y, Takemura K, Tokunaga K, Matsuzawa Y. Enhanced expression of PAI-1 in visceral fat: possible contributor to vascular disease in obesity. Nat Med. 1996;2:800–803. doi: 10.1038/nm0796-800. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

