Multiscale methods for macromolecular simulations
- PMID: 18721882
- PMCID: PMC6407689
- DOI: 10.1016/j.sbi.2008.07.003
Multiscale methods for macromolecular simulations
Abstract
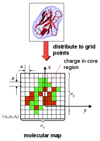
In this article we review the key modeling tools available for simulating biomolecular systems. We consider recent developments and representative applications of mixed quantum mechanics/molecular mechanics (QM/MM), elastic network models (ENMs), coarse-grained molecular dynamics, and grid-based tools for calculating interactions between essentially rigid protein assemblies. We consider how the different length scales can be coupled, both in a sequential fashion (e.g. a coarse-grained or grid model using parameterization from MD simulations), and via concurrent approaches, where the calculations are performed together and together control the progression of the simulation. We suggest how the concurrent coupling approach familiar in the context of QM/MM calculations can be generalized, and describe how this has been done in the CHARMM macromolecular simulation package.
Figures

References
-
- Schulten K, Phillips JC, Kale LV, Bhatele A. Biomolecular modeling in the era of petascale computing. In: Bader D, editor. Petascale Computing: Algorithms and Applications. Chapman & Hall / CRC Press; 2008. pp. 165–181.
-
- Tozzini V. Coarse-grained models for proteins. Curr Opin Struct Biol. 2005;15:144–50. - PubMed
-
- Ayton GS, Noid WG, Voth GA. Multiscale modeling of biomolecular systems: in serial and in parallel. Current Opinion in Structural Biology. 2007;17:192–198. - PubMed
-
- Lu G, Kaxiras E. An Overview of Multiscale Simulations of Materials. In: Rieth M, Schommers W, editors. Handbook of Theoretical and Computational Nanothechnology. X. American Scientific Publishers; 2005. pp. 1–33.
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- BBS/B/16011/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BEP17032/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- B19456/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

