Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima
- PMID: 18722384
- PMCID: PMC2628389
- DOI: 10.1016/j.jmb.2008.08.012
Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima
Abstract
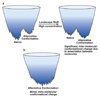
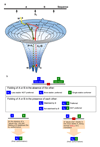
How can we understand a case in which a given amino acid sequence folds into structurally and functionally distinct molecules? Synonymous single-nucleotide polymorphisms in the MDR1 (multidrug resistance 1 or ABCB1) gene involving frequent-to-rare codon substitutions lead to identical protein sequences. Remarkably, these alternative sequences give a protein product with similar but different structures and functions. Here, we propose that long-enough ribosomal pause time scales may lead to alternate folding pathways and distinct minima on the folding free energy surface. While the conformational and functional differences between the native and alternate states may be minor, the MDR1 case illustrates that the barriers may nevertheless constitute sufficiently high hurdles in physiological time scales, leading to kinetically trapped states with altered structures and functions. Different folding pathways leading to conformationally similar trapped states may be due to swapping of (fairly symmetric) segments. Domain swapping is more likely in the no-pause case in which the chain elongates and folds simultaneously; on the other hand, sufficiently long pause times between such segments may be expected to lessen the chances of swapping events. Here, we review the literature in this light.
Figures
References
-
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. - PubMed
-
- Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nature Reviews Drug Discovery. 2006;5:219–234. - PubMed
-
- Markova S, Nakamura T, Sakaeda T, Makimoto H, Uchiyama H, Okamura N, Okumura K. Genotype-dependent down-regulation of gene expression and function of MDR1 in human peripheral blood mononuclear cells under acute inflammation. Drug Metab Pharmacokinet. 2006;21:194–200. - PubMed
-
- Asano T, Takahashi KA, Fujioka M, Inoue S, Okamoto M, Sugioka N, Nishino H, Tanaka T, Hirota Y, Kubo T. ABCB1 C3435T and G2677T/A polymorphism decreased the risk for steroid-induced osteonecrosis of the femoral head after kidney transplantation. Pharmacogenetics. 2003;13:675–682. - PubMed
-
- Tan EK, Chan DKY, Ng PW, Woo J, Teo YY, Tang K, Wong LP, Chong SS, Tan C, Shen H, Zhao Y, Lee CGL. Effect of MDR1 haplotype on risk of Parkinson disease. Arch. Neurol. 2005;62:460–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

