A conserved Cys-loop receptor aspartate residue in the M3-M4 cytoplasmic loop is required for GABAA receptor assembly
- PMID: 18723504
- PMCID: PMC2573071
- DOI: 10.1074/jbc.M802856200
A conserved Cys-loop receptor aspartate residue in the M3-M4 cytoplasmic loop is required for GABAA receptor assembly
Abstract
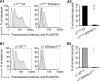
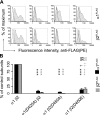
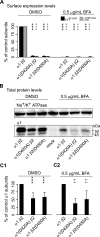
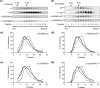
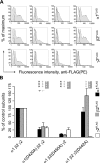
Members of the Cys-loop superfamily of ligand-gated ion channels, which mediate fast synaptic transmission in the nervous system, are assembled as heteropentamers from a large repertoire of neuronal subunits. Although several motifs in subunit N-terminal domains are known to be important for subunit assembly, increasing evidence points toward a role for C-terminal domains. Using a combination of flow cytometry, patch clamp recording, endoglycosidase H digestion, brefeldin A treatment, and analytic centrifugation, we identified a highly conserved aspartate residue at the boundary of the M3-M4 loop and the M4 domain that was required for binary and ternary gamma-aminobutyric acid type A receptor surface expression. Mutation of this residue caused mutant and partnering subunits to be retained in the endoplasmic reticulum, reflecting impaired forward trafficking. Interestingly although mutant and partnering wild type subunits could be coimmunoprecipitated, analytic centrifugation studies demonstrated decreased formation of pentameric receptors, suggesting that this residue played an important role in later steps of subunit oligomerization. We thus conclude that C-terminal motifs are also important determinants of Cys-loop receptor assembly.
Figures






Similar articles
-
Glycosylation of {beta}2 subunits regulates GABAA receptor biogenesis and channel gating.J Biol Chem. 2010 Oct 8;285(41):31348-61. doi: 10.1074/jbc.M110.151449. Epub 2010 Jul 16. J Biol Chem. 2010. PMID: 20639197 Free PMC article.
-
The minimum M3-M4 loop length of neurotransmitter-activated pentameric receptors is critical for the structural integrity of cytoplasmic portals.J Biol Chem. 2013 Jul 26;288(30):21558-68. doi: 10.1074/jbc.M113.481689. Epub 2013 Jun 5. J Biol Chem. 2013. PMID: 23740249 Free PMC article.
-
A conserved cysteine residue in the third transmembrane domain is essential for homomeric 5-HT3 receptor function.J Physiol. 2010 Feb 15;588(Pt 4):603-16. doi: 10.1113/jphysiol.2009.181719. Epub 2009 Nov 23. J Physiol. 2010. PMID: 19933756 Free PMC article.
-
Regulation of ligand-gated ion channels by protein phosphorylation.Adv Second Messenger Phosphoprotein Res. 1999;33:49-78. doi: 10.1016/s1040-7952(99)80005-6. Adv Second Messenger Phosphoprotein Res. 1999. PMID: 10218114 Review.
-
Assembly of nicotinic and other Cys-loop receptors.J Neurochem. 2011 Mar;116(5):734-41. doi: 10.1111/j.1471-4159.2010.07060.x. Epub 2011 Jan 13. J Neurochem. 2011. PMID: 21214570 Review.
Cited by
-
Glycosylation of {beta}2 subunits regulates GABAA receptor biogenesis and channel gating.J Biol Chem. 2010 Oct 8;285(41):31348-61. doi: 10.1074/jbc.M110.151449. Epub 2010 Jul 16. J Biol Chem. 2010. PMID: 20639197 Free PMC article.
-
Slow degradation and aggregation in vitro of mutant GABAA receptor gamma2(Q351X) subunits associated with epilepsy.J Neurosci. 2010 Oct 13;30(41):13895-905. doi: 10.1523/JNEUROSCI.2320-10.2010. J Neurosci. 2010. PMID: 20943930 Free PMC article.
-
Impaired surface αβγ GABA(A) receptor expression in familial epilepsy due to a GABRG2 frameshift mutation.Neurobiol Dis. 2013 Feb;50:135-41. doi: 10.1016/j.nbd.2012.10.008. Epub 2012 Oct 13. Neurobiol Dis. 2013. PMID: 23069679 Free PMC article.
-
An intracellular redox sensor for reactive oxygen species at the M3-M4 linker of GABAA ρ1 receptors.Br J Pharmacol. 2014 May;171(9):2291-9. doi: 10.1111/bph.12581. Br J Pharmacol. 2014. PMID: 24428763 Free PMC article.
-
Deorphanization of novel biogenic amine-gated ion channels identifies a new serotonin receptor for learning.Curr Biol. 2021 Oct 11;31(19):4282-4292.e6. doi: 10.1016/j.cub.2021.07.036. Epub 2021 Aug 12. Curr Biol. 2021. PMID: 34388373 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

