Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development
- PMID: 18723577
- PMCID: PMC2553617
- DOI: 10.1105/tpc.108.059584
Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development
Abstract
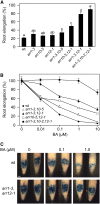
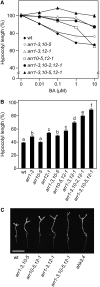
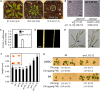
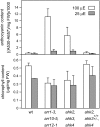
The type B Arabidopsis Response Regulators (ARRs) of Arabidopsis thaliana are transcription factors that act as positive regulators in the two-component cytokinin signaling pathway. We employed a mutant-based approach to perform a detailed characterization of the roles of ARR1, ARR10, and ARR12 in plant growth and development. The most pronounced phenotype was found in the arr1-3 arr10-5 arr12-1 triple loss-of-function mutant, which showed almost complete insensitivity to high levels of exogenously applied cytokinins. The triple mutant exhibited reduced stature due to decreased cell division in the shoot, enhanced seed size, increased sensitivity to light, altered chlorophyll and anthocyanin concentrations, and an aborted primary root with protoxylem but no metaxylem. Microarray analysis revealed that expression of the majority of cytokinin-regulated genes requires the function of ARR1, ARR10, and ARR12. Characterization of double mutants revealed differing contributions of the type B ARRs to mutant phenotypes. Our results support a model in which cytokinin regulates a wide array of downstream responses through the action of a multistep phosphorelay that culminates in transcriptional regulation by ARR1, ARR10, and ARR12.
Figures



References
-
- Bianchi, M.W., Roux, C., and Vartanian, N. (2002). Drought regulation of GST8, encoding the Arabidopsis homologue of ParC/Nt107 glutathione transferase/peroxidase. Physiol. Plant. 116 96–105. - PubMed
-
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. - PubMed
-
- Brenner, W.G., Romanov, G.A., Kollmer, I., Burkle, L., and Schmulling, T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44 314–333. - PubMed
-
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance. Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

