Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe
- PMID: 18723604
- PMCID: PMC2568073
- DOI: 10.1128/EC.00230-08
Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe
Abstract
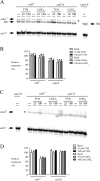
Copper amine oxidases (CAOs) have been proposed to be involved in the metabolism of xenobiotic and biogenic amines. The requirement for copper is absolute for their activity. In the fission yeast Schizosaccharomyces pombe, cao1(+) and cao2(+) genes are predicted to encode members of the CAO family. While both genes are expressed in wild-type cells, we determined that the expression of only cao1(+) but not cao2(+) results in the production of an active enzyme. Site-directed mutagenesis identified three histidine residues within the C-terminal region of Cao1 that are necessary for amine oxidase activity. By use of a cao1(+)-GFP allele that retained wild-type function, Cao1-GFP was localized in the cytosol (GFP is green fluorescent protein). Under copper-limiting conditions, disruption of ctr4(+), ctr5(+), and cuf1(+) produced a defect in amine oxidase activity, indicating that a functionally active Cao1 requires Ctr4/5-mediated copper transport and the transcription factor Cuf1. Likewise, atx1 null cells exhibited substantially decreased levels of amine oxidase activity. In contrast, deletion of ccc2, cox17, and pccs had no significant effect on Cao1 activity. Residual amine oxidase activity in cells lacking atx1(+) can be restored to normal levels by returning an atx1(+) allele, underscoring the critical importance of the presence of Atx1 in cells. Using two-hybrid analysis, we demonstrated that Cao1 physically interacts with Atx1 and that this association is comparable to that of Atx1 with the N-terminal region of Ccc2. Collectively, these results describe the first example of the ability of Atx1 to act as a copper carrier for a molecule other than Ccc2 and its critical role in delivering copper to Cao1.
Figures





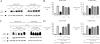


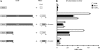
Similar articles
-
Characterization of Schizosaccharomyces pombe copper transporter proteins in meiotic and sporulating cells.J Biol Chem. 2014 Apr 4;289(14):10168-81. doi: 10.1074/jbc.M113.543678. Epub 2014 Feb 25. J Biol Chem. 2014. PMID: 24569997 Free PMC article.
-
Mechanisms of copper loading on the Schizosaccharomyces pombe copper amine oxidase 1 expressed in Saccharomyces cerevisiae.Microbiology (Reading). 2006 Sep;152(Pt 9):2819-2830. doi: 10.1099/mic.0.28998-0. Microbiology (Reading). 2006. PMID: 16946276
-
Cell-surface copper transporters and superoxide dismutase 1 are essential for outgrowth during fungal spore germination.J Biol Chem. 2017 Jul 14;292(28):11896-11914. doi: 10.1074/jbc.M117.794677. Epub 2017 Jun 1. J Biol Chem. 2017. PMID: 28572514 Free PMC article.
-
Copper transport and regulation in Schizosaccharomyces pombe.Biochem Soc Trans. 2013 Dec;41(6):1679-86. doi: 10.1042/BST2013089. Biochem Soc Trans. 2013. PMID: 24256274 Free PMC article. Review.
-
[The molecular bases for copper uptake and distribution: lessons from yeast].Med Sci (Paris). 2008 Mar;24(3):277-83. doi: 10.1051/medsci/2008243277. Med Sci (Paris). 2008. PMID: 18334176 Review. French.
Cited by
-
Copper-dependent trafficking of the Ctr4-Ctr5 copper transporting complex.PLoS One. 2010 Aug 4;5(8):e11964. doi: 10.1371/journal.pone.0011964. PLoS One. 2010. PMID: 20694150 Free PMC article.
-
Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells.J Biol Chem. 2011 Sep 30;286(39):34356-72. doi: 10.1074/jbc.M111.280396. Epub 2011 Aug 2. J Biol Chem. 2011. PMID: 21828039 Free PMC article.
-
Copper Acquisition and Utilization in Fungi.Annu Rev Microbiol. 2017 Sep 8;71:597-623. doi: 10.1146/annurev-micro-030117-020444. Annu Rev Microbiol. 2017. PMID: 28886682 Free PMC article. Review.
-
abc3+ encodes an iron-regulated vacuolar ABC-type transporter in Schizosaccharomyces pombe.Eukaryot Cell. 2010 Jan;9(1):59-73. doi: 10.1128/EC.00262-09. Epub 2009 Nov 13. Eukaryot Cell. 2010. PMID: 19915076 Free PMC article.
-
Characterization of Schizosaccharomyces pombe copper transporter proteins in meiotic and sporulating cells.J Biol Chem. 2014 Apr 4;289(14):10168-81. doi: 10.1074/jbc.M113.543678. Epub 2014 Feb 25. J Biol Chem. 2014. PMID: 24569997 Free PMC article.
References
-
- Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast, a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Beaudoin, J., and S. Labbé. 2001. The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an Ace1/Amt1-like recognition sequence. J. Biol. Chem. 27615472-15480. - PubMed
-
- Beaudoin, J., J. Laliberté, and S. Labbé. 2006. Functional dissection of Ctr4 and Ctr5 amino-terminal regions reveals motifs with redundant roles in copper transport. Microbiology 152209-222. - PubMed
-
- Beaudoin, J., A. Mercier, R. Langlois, and S. Labbé. 2003. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 27814565-14577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

