The origins of 168, W23, and other Bacillus subtilis legacy strains
- PMID: 18723616
- PMCID: PMC2580678
- DOI: 10.1128/JB.00722-08
The origins of 168, W23, and other Bacillus subtilis legacy strains
Abstract
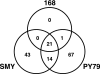
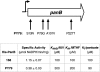
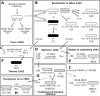
Bacillus subtilis is both a model organism for basic research and an industrial workhorse, yet there are major gaps in our understanding of the genomic heritage and provenance of many widely used strains. We analyzed 17 legacy strains dating to the early years of B. subtilis genetics. For three--NCIB 3610T, PY79, and SMY--we performed comparative genome sequencing. For the remainder, we used conventional sequencing to sample genomic regions expected to show sequence heterogeneity. Sequence comparisons showed that 168, its siblings (122, 160, and 166), and the type strains NCIB 3610 and ATCC 6051 are highly similar and are likely descendants of the original Marburg strain, although the 168 lineage shows genetic evidence of early domestication. Strains 23, W23, and W23SR are identical in sequence to each other but only 94.6% identical to the Marburg group in the sequenced regions. Strain 23, the probable W23 parent, likely arose from a contaminant in the mutagenesis experiments that produced 168. The remaining strains are all genomic hybrids, showing one or more "W23 islands" in a 168 genomic backbone. Each traces its origin to transformations of 168 derivatives with DNA from 23 or W23. The common prototrophic lab strain PY79 possesses substantial W23 islands at its trp and sac loci, along with large deletions that have reduced its genome 4.3%. SMY, reputed to be the parent of 168, is actually a 168-W23 hybrid that likely shares a recent ancestor with PY79. These data provide greater insight into the genomic history of these B. subtilis legacy strains.
Figures


References
-
- Aertsen, A., I. Van Opstal, S. C. Vanmuysen, E. Y. Wuytack, and C. W. Michiels. 2005. Screening for Bacillus subtilis mutants deficient in pressure induced spore germination: identification of ykvU as a novel germination gene. FEMS Microbiol. Lett. 243385-391. - PubMed
-
- Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2951-953. - PubMed
-
- Albertini, A. M., and A. Galizzi. 1999. The sequence of the trp operon of Bacillus subtilis 168 (trpC2) revisited. Microbiology 1453319-3320. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

