Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA
- PMID: 18723618
- PMCID: PMC2580700
- DOI: 10.1128/JB.00676-08
Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA
Abstract
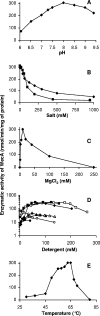
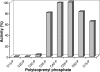
To date, the structural and functional characterization of proteins belonging to the polyprenyl-phosphate N-acetylhexosamine-1-phosphate transferase superfamily has been relentlessly held back by problems encountered with their overexpression and purification. In the present work and for the first time, the integral membrane protein WecA that catalyzes the transfer of the GlcNAc-1-phosphate moiety from UDP-GlcNAc onto the carrier lipid undecaprenyl phosphate, yielding undecaprenyl-pyrophosphoryl-GlcNAc, the lipid intermediate involved in the synthesis of various bacterial cell envelope components, was overproduced and purified to near homogeneity in milligram quantities. An enzymatic assay was developed, and the kinetic parameters of WecA as well as the effects of pH, salts, cations, detergents, and temperature on the enzyme activity were determined. A minimal length of 35 carbons was required for the lipid substrate, and tunicamycin was shown to inhibit the enzyme at submicromolar concentrations.
Figures


Similar articles
-
Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide.J Bacteriol. 2007 Apr;189(7):2618-28. doi: 10.1128/JB.01905-06. Epub 2007 Jan 19. J Bacteriol. 2007. PMID: 17237164 Free PMC article.
-
Conserved amino acid residues found in a predicted cytosolic domain of the lipopolysaccharide biosynthetic protein WecA are implicated in the recognition of UDP-N-acetylglucosamine.Microbiology (Reading). 2001 Nov;147(Pt 11):3015-25. doi: 10.1099/00221287-147-11-3015. Microbiology (Reading). 2001. PMID: 11700352
-
Preparative enzymatic synthesis of polyprenyl-pyrophosphoryl-N-acetylglucosamine, an essential lipid intermediate for the biosynthesis of various bacterial cell envelope polymers.Anal Biochem. 2009 Aug 15;391(2):163-5. doi: 10.1016/j.ab.2009.05.011. Epub 2009 May 12. Anal Biochem. 2009. PMID: 19442646
-
Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases.Glycobiology. 2005 Sep;15(9):29R-42R. doi: 10.1093/glycob/cwi065. Epub 2005 Apr 20. Glycobiology. 2005. PMID: 15843595 Review.
-
Structural basis for selective inhibition of antibacterial target MraY, a membrane-bound enzyme involved in peptidoglycan synthesis.Drug Discov Today. 2018 Jul;23(7):1426-1435. doi: 10.1016/j.drudis.2018.05.020. Epub 2018 May 18. Drug Discov Today. 2018. PMID: 29778697 Review.
Cited by
-
CbrA Mediates Colicin M Resistance in Escherichia coli through Modification of Undecaprenyl-Phosphate-Linked Peptidoglycan Precursors.J Bacteriol. 2020 Nov 4;202(23):e00436-20. doi: 10.1128/JB.00436-20. Print 2020 Nov 4. J Bacteriol. 2020. PMID: 32958631 Free PMC article.
-
WGS-Based Lineage and Antimicrobial Resistance Pattern of Salmonella Typhimurium Isolated during 2000-2017 in Peru.Antibiotics (Basel). 2022 Aug 30;11(9):1170. doi: 10.3390/antibiotics11091170. Antibiotics (Basel). 2022. PMID: 36139949 Free PMC article.
-
Subcellular localization of the enterobacterial common antigen GT-E-like glycosyltransferase, WecG.Mol Microbiol. 2022 Oct;118(4):403-416. doi: 10.1111/mmi.14973. Epub 2022 Aug 25. Mol Microbiol. 2022. PMID: 36006410 Free PMC article.
-
Fluorescence-based assay for polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA) and identification of novel antimycobacterial WecA inhibitors.Anal Biochem. 2016 Nov 1;512:78-90. doi: 10.1016/j.ab.2016.08.008. Epub 2016 Aug 13. Anal Biochem. 2016. PMID: 27530653 Free PMC article.
-
Preference of Bacterial Rhamnosyltransferases for 6-Deoxysugars Reveals a Strategy To Deplete O-Antigens.J Am Chem Soc. 2023 Jul 26;145(29):15639-15646. doi: 10.1021/jacs.3c03005. Epub 2023 Jul 12. J Am Chem Soc. 2023. PMID: 37437030 Free PMC article.
References
-
- Amer, A. O., and M. A. Valvano. 2001. Conserved amino acid residues found in a predicted cytosolic domain of the lipopolysaccharide biosynthetic protein WecA are implicated in the recognition of UDP-N-acetylglucosamine. Microbiology 1473015-3025. - PubMed
-
- Amer, A. O., and M. A. Valvano. 2002. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148571-582. - PubMed
-
- Anderson, M. S., S. S. Eveland, and N. P. Price. 2000. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol. Lett. 191169-175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

