SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus
- PMID: 18723629
- PMCID: PMC2566184
- DOI: 10.1128/JB.00700-08
SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus
Abstract
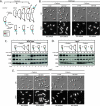
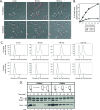
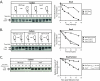
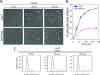
Cell cycle progression and polar differentiation are temporally coordinated in Caulobacter crescentus. This oligotrophic bacterium divides asymmetrically to produce a motile swarmer cell that represses DNA replication and a sessile stalked cell that replicates its DNA. The initiation of DNA replication coincides with the proteolysis of the CtrA replication inhibitor and the accumulation of DnaA, the replication initiator, upon differentiation of the swarmer cell into a stalked cell. We analyzed the adaptive response of C. crescentus swarmer cells to carbon starvation and found that there was a block in both the swarmer-to-stalked cell polar differentiation program and the initiation of DNA replication. SpoT is a bifunctional synthase/hydrolase that controls the steady-state level of the stress-signaling nucleotide (p)ppGpp, and carbon starvation caused a SpoT-dependent increase in (p)ppGpp concentration. Carbon starvation activates DnaA proteolysis (B. Gorbatyuk and G. T. Marczynski, Mol. Microbiol. 55:1233-1245, 2005). We observed that SpoT is required for this phenomenon in swarmer cells, and in the absence of SpoT, carbon-starved swarmer cells inappropriately initiated DNA replication. Since SpoT controls (p)ppGpp abundance, we propose that this nucleotide relays carbon starvation signals to the cellular factors responsible for activating DnaA proteolysis, thereby inhibiting the initiation of DNA replication. SpoT, however, was not required for the carbon starvation block of the swarmer-to-stalked cell polar differentiation program. Thus, swarmer cells utilize at least two independent signaling pathways to relay carbon starvation signals: a SpoT-dependent pathway mediating the inhibition of DNA replication initiation, and a SpoT-independent pathway(s) that blocks morphological differentiation.
Figures


Similar articles
-
ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus.J Bacteriol. 2012 Jan;194(1):28-35. doi: 10.1128/JB.05932-11. Epub 2011 Oct 21. J Bacteriol. 2012. PMID: 22020649 Free PMC article.
-
Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus.J Bacteriol. 2014 Jul;196(14):2514-25. doi: 10.1128/JB.01575-14. Epub 2014 May 2. J Bacteriol. 2014. PMID: 24794566 Free PMC article.
-
Phosphate starvation decouples cell differentiation from DNA replication control in the dimorphic bacterium Caulobacter crescentus.PLoS Genet. 2023 Nov 27;19(11):e1010882. doi: 10.1371/journal.pgen.1010882. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 38011258 Free PMC article.
-
Regulation of chromosomal replication in Caulobacter crescentus.Plasmid. 2012 Mar;67(2):76-87. doi: 10.1016/j.plasmid.2011.12.007. Epub 2011 Dec 29. Plasmid. 2012. PMID: 22227374 Review.
-
Multilayered control of chromosome replication in Caulobacter crescentus.Biochem Soc Trans. 2019 Feb 28;47(1):187-196. doi: 10.1042/BST20180460. Epub 2019 Jan 9. Biochem Soc Trans. 2019. PMID: 30626709 Free PMC article. Review.
Cited by
-
Topological control of the Caulobacter cell cycle circuitry by a polarized single-domain PAS protein.Nat Commun. 2015 May 8;6:7005. doi: 10.1038/ncomms8005. Nat Commun. 2015. PMID: 25952018 Free PMC article.
-
ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus.J Bacteriol. 2012 Jan;194(1):28-35. doi: 10.1128/JB.05932-11. Epub 2011 Oct 21. J Bacteriol. 2012. PMID: 22020649 Free PMC article.
-
Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus.PLoS One. 2011;6(10):e26028. doi: 10.1371/journal.pone.0026028. Epub 2011 Oct 14. PLoS One. 2011. PMID: 22022497 Free PMC article.
-
The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis.J Bacteriol. 2009 Jun;191(11):3736-9. doi: 10.1128/JB.00216-09. Epub 2009 Mar 27. J Bacteriol. 2009. PMID: 19329632 Free PMC article.
-
Replication initiator DnaA binds at the Caulobacter centromere and enables chromosome segregation.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):16100-5. doi: 10.1073/pnas.1418989111. Epub 2014 Oct 27. Proc Natl Acad Sci U S A. 2014. PMID: 25349407 Free PMC article.
References
-
- Alley, M. R., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science 2591754-1757. - PubMed
-
- Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305673-688. - PubMed
-
- Battesti, A., and E. Bouveret. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 621048-1063. - PubMed
-
- Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 101581-1592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

