Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing
- PMID: 18723673
- PMCID: PMC2529122
- DOI: 10.1073/pnas.0801523105
Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing
Abstract
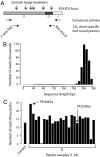
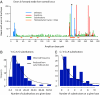
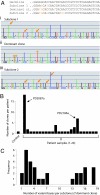
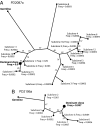
During the clonal expansion of cancer from an ancestral cell with an initiating oncogenic mutation to symptomatic neoplasm, the occurrence of somatic mutations (both driver and passenger) can be used to track the on-going evolution of the neoplasm. All subclones within a cancer are phylogenetically related, with the prevalence of each subclone determined by its evolutionary fitness and the timing of its origin relative to other subclones. Recently developed massively parallel sequencing platforms promise the ability to detect rare subclones of genetic variants without a priori knowledge of the mutations involved. We used ultra-deep pyrosequencing to investigate intraclonal diversification at the Ig heavy chain locus in 22 patients with B-cell chronic lymphocytic leukemia. Analysis of a non-polymorphic control locus revealed artifactual insertions and deletions resulting from sequencing errors and base substitutions caused by polymerase misincorporation during PCR amplification. We developed an algorithm to differentiate genuine haplotypes of somatic hypermutations from such artifacts. This proved capable of detecting multiple rare subclones with frequencies as low as 1 in 5000 copies and allowed the characterization of phylogenetic interrelationships among subclones within each patient. This study demonstrates the potential for ultra-deep resequencing to recapitulate the dynamics of clonal evolution in cancer cell populations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. - PubMed
-
- Davies H, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. - PubMed
-
- Roche-Lestienne C, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. - PubMed
-
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukemia. Nat Rev Cancer. 2003;3:639–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

