Regulatory networks define phenotypic classes of human stem cell lines
- PMID: 18724358
- PMCID: PMC2637443
- DOI: 10.1038/nature07213
Regulatory networks define phenotypic classes of human stem cell lines
Abstract
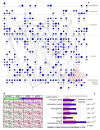
Stem cells are defined as self-renewing cell populations that can differentiate into multiple distinct cell types. However, hundreds of different human cell lines from embryonic, fetal and adult sources have been called stem cells, even though they range from pluripotent cells-typified by embryonic stem cells, which are capable of virtually unlimited proliferation and differentiation-to adult stem cell lines, which can generate a far more limited repertoire of differentiated cell types. The rapid increase in reports of new sources of stem cells and their anticipated value to regenerative medicine has highlighted the need for a general, reproducible method for classification of these cells. We report here the creation and analysis of a database of global gene expression profiles (which we call the 'stem cell matrix') that enables the classification of cultured human stem cells in the context of a wide variety of pluripotent, multipotent and differentiated cell types. Using an unsupervised clustering method to categorize a collection of approximately 150 cell samples, we discovered that pluripotent stem cell lines group together, whereas other cell types, including brain-derived neural stem cell lines, are very diverse. Using further bioinformatic analysis we uncovered a protein-protein network (PluriNet) that is shared by the pluripotent cells (embryonic stem cells, embryonal carcinomas and induced pluripotent cells). Analysis of published data showed that the PluriNet seems to be a common characteristic of pluripotent cells, including mouse embryonic stem and induced pluripotent cells and human oocytes. Our results offer a new strategy for classifying stem cells and support the idea that pluripotency and self-renewal are under tight control by specific molecular networks.
Figures


References
-
- Müller FJ, Snyder EY, Loring JF. Gene therapy: can neural stem cells deliver? Nat Rev Neurosci. 2006;7:75–84. - PubMed
-
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–80. - PubMed
-
- Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–16. - PubMed
-
- Gao Y, Church G. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics. 2005;21:3970–5. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

