Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter
- PMID: 18725401
- PMCID: PMC2573233
- DOI: 10.1128/MCB.00398-08
Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter
Abstract
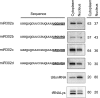
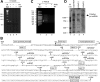
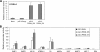
MicroRNAs (miRNAs) play a central role in the regulation of multiple biological processes including the maintenance of stem cell self-renewal and pluripotency. Recently, the miRNA cluster miR302-367 was shown to be differentially expressed in embryonic stem cells (ESCs). Unfortunately, very little is known about the genomic structure of miRNA-encoding genes and their transcriptional units. Here, we have characterized the structure of the gene coding for the human miR302-367 cluster. We identify the transcriptional start and functional core promoter region which specifically drives the expression of this miRNA cluster. The promoter activity depends on the ontogeny and hierarchical cellular stage. It is functional during embryonic development, but it is turned off later in development. From a hierarchical standpoint, its activity decays upon differentiation of ESCs, suggesting that its activity is restricted to the ESC compartment and that the ESC-specific expression of the miR302-367 cluster is fully conferred by its core promoter transcriptional activity. Furthermore, algorithmic prediction of transcription factor binding sites and knockdown studies suggest that ESC-associated transcription factors, including Nanog, Oct3/4, Sox2, and Rex1 may be upstream regulators of miR302-367 promoter. This study represents the first identification, characterization, and functional validation of a human miRNA promoter in stem cells. This study opens up new avenues to further investigate the upstream transcriptional regulation of the miR302-367 cluster and to dissect how these miRNAs integrate in the complex molecular network conferring stem cell properties to ESCs.
Figures



References
-
- Ambros, V. 2004. The functions of animal miRNAs. Nature 435350-355. - PubMed
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. - PubMed
-
- Bernstein, E., S. Y. Kim, M. A. Carmell, E. P. Murchison, H. Alcorn, M. Z. Li, A. A. Mills, S. J. Elledge, K. V. Anderson, and G. J. Hannon. 2003. Dicer is essential for mouse development. Nat. Genet. 35215-217. - PubMed
-
- Borchert, G., W. Lanier, and B. Davidson. 2006. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 131097-1101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
