Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst
- PMID: 18725515
- PMCID: PMC2768606
- DOI: 10.1242/dev.021519
Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst
Abstract
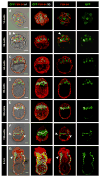
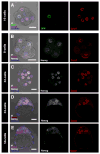
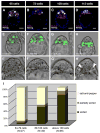
The first two lineages to differentiate from a pluripotent cell population during mammalian development are the extraembryonic trophectoderm (TE) and the primitive endoderm (PrE). Whereas the mechanisms of TE specification have been extensively studied, segregation of PrE and the pluripotent epiblast (EPI) has received comparatively little attention. A current model of PrE specification suggests PrE precursors exhibit an apparently random distribution within the inner cell mass of the early blastocyst and then segregate to their final position lining the cavity by the late blastocyst. We have identified platelet-derived growth factor receptor alpha (Pdgfralpha) as an early-expressed protein that is also a marker of the later PrE lineage. By combining live imaging of embryos expressing a histone H2B-GFP fusion protein reporter under the control of Pdgfra regulatory elements with the analysis of lineage-specific markers, we investigated the events leading to PrE and EPI lineage segregation in the mouse, and correlated our findings using an embryo staging system based on total cell number. Before blastocyst formation, lineage-specific factors are expressed in an overlapping manner. Subsequently, a gradual progression towards a mutually exclusive expression of PrE- and EPI-specific markers occurs. Finally, cell sorting is achieved by a variety of cell behaviours and by selective apoptosis.
Figures




References
-
- Becker S, Casanova J, Grabel L. Localization of endoderm-specific mRNAs in differentiating F9 embryoid bodies. Mech Dev. 1992;37:3–12. - PubMed
-
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. - PubMed
-
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. - PubMed
-
- Copp AJ. Interaction between inner cell mass and trophectoderm of the mouse blastocyst. I A study of cellular proliferation. J Embryol Exp Morphol. 1978;48:109–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

