SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34
- PMID: 18725540
- PMCID: PMC2518712
- DOI: 10.1083/jcb.200804131
SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34
Abstract
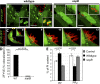
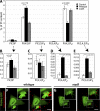
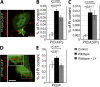
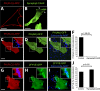
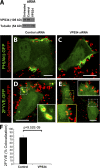
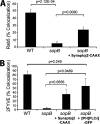
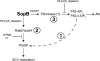
Salmonella colonizes a vacuolar niche in host cells during infection. Maturation of the Salmonella-containing vacuole (SCV) involves the formation of phosphatidylinositol 3-phosphate (PI(3)P) on its outer leaflet. SopB, a bacterial virulence factor with phosphoinositide phosphatase activity, was proposed to generate PI(3)P by dephosphorylating PI(3,4)P2, PI(3,5)P2, and PI(3,4,5)P3. Here, we examine the mechanism of PI(3)P formation during Salmonella infection. SopB is required to form PI(3,4)P2/PI(3,4,5)P3 at invasion ruffles and PI(3)P on nascent SCVs. However, we uncouple these events experimentally and reveal that SopB does not dephosphorylate PI(3,4)P2/PI(3,4,5)P3 to produce PI(3)P. Instead, the phosphatase activity of SopB is required for Rab5 recruitment to the SCV. Vps34, a PI3-kinase that associates with active Rab5, is responsible for PI(3)P formation on SCVs. Therefore, SopB mediates PI(3)P production on the SCV indirectly through recruitment of Rab5 and its effector Vps34. These findings reveal a link between phosphoinositide phosphatase activity and the recruitment of Rab5 to phagosomes.
Figures


References
-
- Anderson, K.E., and S.P. Jackson. 2003. Class I phosphoinositide 3-kinases. Int. J. Biochem. Cell Biol. 35:1028–1033. - PubMed
-
- Baldeon, M.E., B.P. Ceresa, and J.E. Casanova. 2001. Expression of constitutively active Rab5 uncouples maturation of the Salmonella-containing vacuole from intracellular replication. Cell. Microbiol. 3:473–486. - PubMed
-
- Birmingham, C.L., A.C. Smith, M.A. Bakowski, T. Yoshimori, and J.H. Brumell. 2006. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281:11374–11383. - PubMed
-
- Brumell, J.H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78–84. - PubMed
-
- Bucci, C., A. Lutcke, O. Steele-Mortimer, V.M. Olkkonen, P. Dupree, M. Chiariello, C.B. Bruni, K. Simons, and M. Zerial. 1995. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 366:65–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

