Increasing cloning possibilities using artificial zinc finger nucleases
- PMID: 18725642
- PMCID: PMC2529078
- DOI: 10.1073/pnas.0803618105
Increasing cloning possibilities using artificial zinc finger nucleases
Abstract
The ability to accurately digest and ligate DNA molecules of different origins is fundamental to modern recombinant DNA research. Only a handful of enzymes are capable of recognizing and cleaving novel and long DNA sequences, however. The slow evolution and engineering of new restriction enzymes calls for alternative strategies to design novel and unique restriction enzymes capable of binding and digesting specific long DNA sequences. Here we report on the use of zinc finger nucleases (ZFNs)-hybrid synthetic restriction enzymes that can be specifically designed to bind and cleave long DNA sequences-for the purpose of DNA recombination. We show that novel ZFNs can be designed for the digestion of specific sequences and can be expressed and used for cloning purposes. We also demonstrate the power of ZFNs in DNA cloning by custom-cloning a target DNA sequence and assembling dual-expression cassettes on a single target plasmid, a task that rarely can be achieved using type-II restriction enzymes. We demonstrate the flexibility of ZFN design and the ability to shuffle monomers of different ZFNs for the digestion of compatible recognition sites through ligation of compatible ends and their cleavage by heterodimer ZFNs. Of no less importance, we show that ZFNs can be designed to recognize and cleave existing DNA sequences for the custom-cloning of native target DNA molecules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 . (E) Sequence analysis of the ZFN10 ligation site in one of the pSAT10-
. (E) Sequence analysis of the ZFN10 ligation site in one of the pSAT10- plasmids. Sequences of the reconstructed ZFN10 palindrome-like recognition sites are in blue.
plasmids. Sequences of the reconstructed ZFN10 palindrome-like recognition sites are in blue.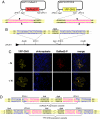


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

