Structural basis for suppression of a host antiviral response by influenza A virus
- PMID: 18725644
- PMCID: PMC2522260
- DOI: 10.1073/pnas.0805213105
Structural basis for suppression of a host antiviral response by influenza A virus
Abstract
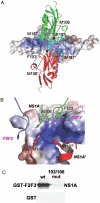
Influenza A viruses are responsible for seasonal epidemics and high mortality pandemics. A major function of the viral NS1A protein, a virulence factor, is the inhibition of the production of IFN-beta mRNA and other antiviral mRNAs. The NS1A protein of the human influenza A/Udorn/72 (Ud) virus inhibits the production of these antiviral mRNAs by binding the cellular 30-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF30), which is required for the 3' end processing of all cellular pre-mRNAs. Here we report the 1.95-A resolution X-ray crystal structure of the complex formed between the second and third zinc finger domain (F2F3) of CPSF30 and the C-terminal domain of the Ud NS1A protein. The complex is a tetramer, in which each of two F2F3 molecules wraps around two NS1A effector domains that interact with each other head-to-head. This structure identifies a CPSF30 binding pocket on NS1A comprised of amino acid residues that are highly conserved among human influenza A viruses. Single amino acid changes within this binding pocket eliminate CPSF30 binding, and a recombinant Ud virus expressing an NS1A protein with such a substitution is attenuated and does not inhibit IFN-beta pre-mRNA processing. This binding pocket is a potential target for antiviral drug development. The crystal structure also reveals that two amino acids outside of this pocket, F103 and M106, which are highly conserved (>99%) among influenza A viruses isolated from humans, participate in key hydrophobic interactions with F2F3 that stabilize the complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hatada E, Takizawa T, Fukuda R. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA. in vitro J Gen Virol. 1992;73:17–25. - PubMed
-
- Chien CY, et al. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. - PubMed
-
- Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

