Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities
- PMID: 18725995
- PMCID: PMC2518522
- DOI: 10.1371/journal.pone.0003042
Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities
Abstract
Background: Sequencing the expressed genetic information of an ecosystem (metatranscriptome) can provide information about the response of organisms to varying environmental conditions. Until recently, metatranscriptomics has been limited to microarray technology and random cloning methodologies. The application of high-throughput sequencing technology is now enabling access to both known and previously unknown transcripts in natural communities.
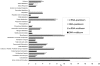
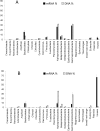
Methodology/principal findings: We present a study of a complex marine metatranscriptome obtained from random whole-community mRNA using the GS-FLX Pyrosequencing technology. Eight samples, four DNA and four mRNA, were processed from two time points in a controlled coastal ocean mesocosm study (Bergen, Norway) involving an induced phytoplankton bloom producing a total of 323,161,989 base pairs. Our study confirms the finding of the first published metatranscriptomic studies of marine and soil environments that metatranscriptomics targets highly expressed sequences which are frequently novel. Our alternative methodology increases the range of experimental options available for conducting such studies and is characterized by an exceptional enrichment of mRNA (99.92%) versus ribosomal RNA. Analysis of corresponding metagenomes confirms much higher levels of assembly in the metatranscriptomic samples and a far higher yield of large gene families with >100 members, approximately 91% of which were novel.
Conclusions/significance: This study provides further evidence that metatranscriptomic studies of natural microbial communities are not only feasible, but when paired with metagenomic data sets, offer an unprecedented opportunity to explore both structure and function of microbial communities--if we can overcome the challenges of elucidating the functions of so many never-seen-before gene families.
Conflict of interest statement
Figures
References
-
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. - PubMed
-
- Handelsman J, Tiedje J, Alvarez-Cohen L, Ashburner M, Cann IKO, et al. The New Science of metagenomics: revealing the secrets of our microbial planet. Washington, DC: The National Academies Press; 2007. - PubMed
-
- Parro V, Moreno-Paz M, Gonzalez-Toril E. Analysis of environmental transcriptomes by DNA microarrays. Env Microbiol. 2007;9:453–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

