Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years
- PMID: 18728074
- PMCID: PMC2727391
- DOI: 10.1093/molbev/msn187
Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years
Abstract
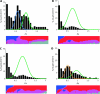
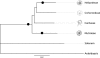
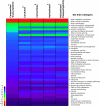
Of the approximately 250,000 species of flowering plants, nearly one in ten are members of the Compositae (Asteraceae), a diverse family found in almost every habitat on all continents except Antarctica. With an origin in the mid Eocene, the Compositae is also a relatively young family with remarkable diversifications during the last 40 My. Previous cytologic and systematic investigations suggested that paleopolyploidy may have occurred in at least one Compositae lineage, but a recent analysis of genomic data was equivocal. We tested for evidence of paleopolyploidy in the evolutionary history of the family using recently available expressed sequence tag (EST) data from the Compositae Genome Project. Combined with data available on GenBank, we analyzed nearly 1 million ESTs from 18 species representing seven genera and four tribes. Our analyses revealed at least three ancient whole-genome duplications in the Compositae-a paleopolyploidization shared by all analyzed taxa and placed near the origin of the family just prior to the rapid radiation of its tribes and independent genome duplications near the base of the tribes Mutisieae and Heliantheae. These results are consistent with previous research implicating paleopolyploidy in the evolution and diversification of the Heliantheae. Further, we observed parallel retention of duplicate genes from the basal Compositae genome duplication across all tribes, despite divergence times of 33-38 My among these lineages. This pattern of retention was also repeated for the paleologs from the Heliantheae duplication. Intriguingly, the categories of genes retained in duplicate were substantially different from those in Arabidopsis. In particular, we found that genes annotated to structural components or cellular organization Gene Ontology categories were significantly enriched among paleologs, whereas genes associated with transcription and other regulatory functions were significantly underrepresented. Our results suggest that paleopolyploidy can yield strikingly consistent signatures of gene retention in plant genomes despite extensive lineage radiations and recurrent genome duplications but that these patterns vary substantially among higher taxonomic categories.
Figures
References
-
- Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005;21:539–543. - PubMed
-
- Aury J, Jaillon O, Duret L, et al. (43 co-authors) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. - PubMed
-
- Baldwin BG, Wessa BL, Panero JL. Nuclear rDNA evidence for major lineages of Helenioid Heliantheae (Compositae) Syst Bot. 2002;27:161–198.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

