Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids
- PMID: 18728184
- PMCID: PMC2525561
- DOI: 10.1073/pnas.0802607105
Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids
Abstract
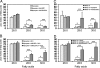
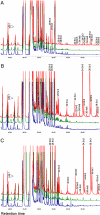
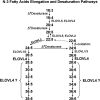
Stargardt-like macular dystrophy (STGD3) is a dominantly inherited juvenile macular degeneration that eventually leads to loss of vision. Three independent mutations causing STGD3 have been identified in exon six of a gene named Elongation of very long chain fatty acids 4 (ELOVL4). The ELOVL4 protein was predicted to be involved in fatty acid elongation, although evidence for this and the specific step(s) it may catalyze have remained elusive. Here, using a gain-of-function approach, we provide direct and compelling evidence that ELOVL4 is required for the synthesis of C28 and C30 saturated fatty acids (VLC-FA) and of C28-C38 very long chain polyunsaturated fatty acids (VLC-PUFA), the latter being uniquely expressed in retina, sperm, and brain. Rat neonatal cardiomyocytes and a human retinal epithelium cell line (ARPE-19) were transduced with recombinant adenovirus type 5 carrying mouse Elovl4 and supplemented with 24:0, 20:5n3, or 22:5n3. The 24:0 was elongated to 28:0 and 30:0; 20:5n3 and 22:5n3 were elongated to a series of C28-C38 PUFA. Because retinal degeneration is the only known phenotype in STGD3 disease, we propose that reduced VLC-PUFA in the retinas of these patients may be the cause of photoreceptor cell death.
Conflict of interest statement
Conflict of interest statement: M.-P.A., R.S.B., and R.E.A. have filed a provisional patent for the use of very long chain polyunsaturated fatty acids in the treatment of macular degeneration.
Figures

References
-
- Zhang K, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93. - PubMed
-
- Edwards AO, Donoso LA, Ritter R., III A novel gene for autosomal dominant Stargardt-like macular dystrophy with homology to the SUR4 protein family. Invest Ophthalmol Vis Sci. 2001;42:2652–2663. - PubMed
-
- Grayson C, Molday RS. Dominant negative mechanism underlies autosomal dominant Stargardt-like macular dystrophy linked to mutations in ELOVL4. J Biol Chem. 2005;280:32521–32530. - PubMed
-
- Ambasudhan R, et al. Atrophic macular degeneration mutations in ELOVL4 result in the intracellular misrouting of the protein. Genomics. 2004;83:615–625. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

