Transcriptional regulation of osteoblasts
- PMID: 18728356
- PMCID: PMC3512205
- DOI: 10.1159/000151747
Transcriptional regulation of osteoblasts
Abstract
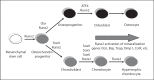
The differentiation of osteoblasts from mesenchymal precursors requires a series of cell fate decisions controlled by a hierarchy of transcription factors. These include RUNX2, Osterix (OSX), ATF4 and a large number of nuclear coregulators. During bone development, initial RUNX2 expression coincides with the formation of mesenchymal condensations and precedes the branching of chondrogenic and osteogenic lineages. Given its central role in bone development, it is not surprising that RUNX2 is subject to a variety of controls. These include posttranslational modification, especially phosphorylation, and interactions with accessory nuclear factors. Specific examples of RUNX2 regulation to be reviewed include phosphorylation by the ERK/MAP kinase pathway and interactions with DLX5. RUNX2 is regulated via phosphorylation of critical serine residues in the proline/serine/threonine domain. In vivo, the transgenic expression of constitutively active MAP kinase in osteoblasts accelerated skeletal development, while a dominant-negative MAPK retarded development in a RUNX2-dependent manner. DLX5-RUNX2 complexes can be detected in osteoblasts and this interaction plays a critical role in maintaining osteoblast-specific expression of the bone sialoprotein gene. These studies allow us to begin understanding the complex mechanisms necessary to fine-tune bone formation as mesenchymal progenitors progress down the osteoblast lineage.
Copyright 2008 S. Karger AG, Basel.
Figures


References
-
- Afzal F., Pratap J., Ito K., Ito Y., Stein J.L., van Wijnen A.J., Stein G.S., Lian J.B., Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:204–63. - PubMed
-
- Benson M.D., Bargeon J.L., Xiao G., Thomas P.E., Kim A., Cui Y., Franceschi R.T. Identification of a homeodomain binding element in the bone sialoprotein gene promoter that is required for its osteoblast-selective expression. J Biol Chem. 2000;275:275–13907. - PubMed
-
- Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., Justice M.J., Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:6–423. - PubMed
-
- Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1 a transcriptional activator of osteoblast differentiation. Cell. 1997;89:89–747. - PubMed
-
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:126–677. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

