Activating transcription factor 4 is critical for proliferation and survival in primary bone marrow stromal cells and calvarial osteoblasts
- PMID: 18729081
- PMCID: PMC2704124
- DOI: 10.1002/jcb.21888
Activating transcription factor 4 is critical for proliferation and survival in primary bone marrow stromal cells and calvarial osteoblasts
Abstract
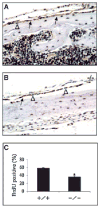
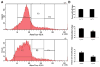
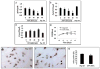
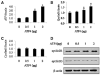
Activating transcription factor 4 (ATF4) is essential for bone formation. However, the mechanism of its actions in bone is poorly understood. The present study examined the role for ATF4 in the regulation of proliferation and survival of primary mouse bone marrow stromal cells (BMSCs) and osteoblasts. Results showed that Atf4(-/-) cells display a severe proliferative defect as measured by multiple cell proliferation assays. Cell cycle progression of Atf4(-/-) BMSCs was largely delayed with significant G1 arrest. Expression of cyclin D1 was decreased both at the mRNA and protein level. A similar proliferation defect was observed in Atf4(-/-) calvarial periosteal osteoblasts when compared with wt control. Knocking down Atf4 mRNA by small interfering RNA in MC3T3-E1 subclone 4 preosteoblasts markedly reduced expression of cyclin D1 and cell proliferation. In contrast, overexpression of ATF4 increased cyclin D1 expression as well as cell proliferation in Atf4(-/-) BMSCs. In addition, apoptosis was significantly increased in Atf4(-/-) BMSCs and calvarial periosteal osteoblasts relative to wt controls. Taken together, these results for the first time demonstrate that ATF4 is a critical regulator of proliferation and survival in BMSCs and osteoblasts in vitro and in vivo.
(c) 2008 Wiley-Liss, Inc.
Figures




References
-
- Adams JM. Ways of dying: Multiple pathways to apoptosis. Genes Dev. 2003;17:2481–2495. - PubMed
-
- Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282:16744–16753. - PubMed
-
- Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL. Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood. 2004;103:1876–1882. - PubMed
-
- Brindle PK, Montminy MR. The CREB family of transcription activators. Curr Opin Genet Dev. 1992;2:199–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

