Molecular characterization of propionyllysines in non-histone proteins
- PMID: 18753126
- PMCID: PMC2621001
- DOI: 10.1074/mcp.M800224-MCP200
Molecular characterization of propionyllysines in non-histone proteins
Abstract
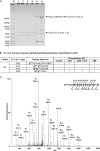
Lysine propionylation and butyrylation are protein modifications that were recently identified in histones. The molecular components involved in the two protein modification pathways are unknown, hindering further functional studies. Here we report identification of the first three in vivo non-histone protein substrates of lysine propionylation in eukaryotic cells: p53, p300, and CREB-binding protein. We used mass spectrometry to map lysine propionylation sites within these three proteins. We also identified the first two in vivo eukaryotic lysine propionyltransferases, p300 and CREB-binding protein, and the first eukaryotic depropionylase, Sirt1. p300 was able to perform autopropionylation on lysine residues in cells. Our results suggest that lysine propionylation, like lysine acetylation, is a dynamic and regulatory post-translational modification. Based on these observations, it appears that some enzymes are common to the lysine propionylation and lysine acetylation regulatory pathways. Our studies therefore identified first several important players in lysine propionylation pathway.
Figures



References
-
- Brownell, J. E., Zhou, J., Ranalli, T., Kobayashi, R., Edmondson, D. G., Roth, S. Y., and Allis, C. D. ( 1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 - PubMed
-
- Gu, W., Shi, X. L., and Roeder, R. G. ( 1997) Synergistic activation of transcription by CBP and p53. Nature 387, 819–823 - PubMed
-
- Taunton, J., Hassig, C. A., and Schreiber, S. L. ( 1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

