The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles
- PMID: 18753196
- PMCID: PMC2573274
- DOI: 10.1128/JVI.01052-08
The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles
Abstract
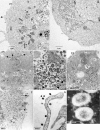
The production of virus-like particles (VLPs) constitutes a relevant and safe model to study molecular determinants of virion egress. The minimal requirement for the assembly of VLPs for the coronavirus responsible for severe acute respiratory syndrome in humans (SARS-CoV) is still controversial. Recent studies have shown that SARS-CoV VLP formation depends on either M and E proteins or M and N proteins. Here we show that both E and N proteins must be coexpressed with M protein for the efficient production and release of VLPs by transfected Vero E6 cells. This suggests that the mechanism of SARS-CoV assembly differs from that of other studied coronaviruses, which only require M and E proteins for VLP formation. When coexpressed, the native envelope trimeric S glycoprotein is incorporated onto VLPs. Interestingly, when a fluorescent protein tag is added to the C-terminal end of N or S protein, but not M protein, the chimeric viral proteins can be assembled within VLPs and allow visualization of VLP production and trafficking in living cells by state-of-the-art imaging technologies. Fluorescent VLPs will be used further to investigate the role of cellular machineries during SARS-CoV egress.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

