Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells
- PMID: 18753283
- PMCID: PMC2556817
- DOI: 10.1104/pp.108.125237
Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells
Abstract
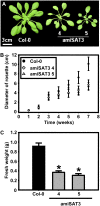
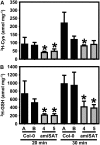
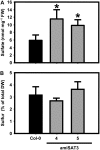
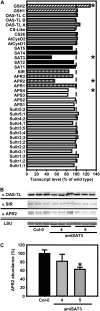
Cysteine (Cys) synthesis in plants is carried out by two sequential reactions catalyzed by the rate-limiting enzyme serine acetyltransferase (SAT) and excess amounts of O-acetylserine(thiol)lyase. Why these reactions occur in plastids, mitochondria, and cytosol of plants remained unclear. Expression of artificial microRNA (amiRNA) against Sat3 encoding mitochondrial SAT3 in transgenic Arabidopsis (Arabidopsis thaliana) plants demonstrates that mitochondria are the most important compartment for the synthesis of O-acetylserine (OAS), the precursor of Cys. Reduction of RNA levels, protein contents, SAT enzymatic activity, and phenotype strongly correlate in independent amiSAT3 lines and cause significantly retarded growth. The expression of the other four Sat genes in the Arabidopsis genome are not affected by amiRNA-SAT3 according to quantitative real-time polymerase chain reaction and microarray analyses. Application of radiolabeled serine to leaf pieces revealed severely reduced incorporation rates into Cys and even more so into glutathione. Accordingly, steady-state levels of OAS are 4-fold reduced. Decrease of sulfate reduction-related genes is accompanied by an accumulation of sulfate in amiSAT3 lines. These results unequivocally show that mitochondria provide the bulk of OAS in the plant cell and are the likely site of flux regulation. Together with recent data, the cytosol appears to be a major site of Cys synthesis, while plastids contribute reduced sulfur as sulfide. Thus, Cys synthesis in plants is significantly different from that in nonphotosynthetic eukaryotes at the cellular level.
Figures



References
-
- Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R (2002) Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J Biol Chem 277 30629–30634 - PubMed
-
- Blaszczyk A, Brodzik R, Sirko A (1999) Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J 20 237–243 - PubMed
-
- Bogdanova N, Bork C, Hell R (1995) Cysteine biosynthesis in plants: isolation and functional identification of a cDNA encoding a serine acetyltransferase from Arabidopsis thaliana. FEBS Lett 358 43–47 - PubMed
-
- Bogdanova N, Hell R (1997) Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J 11 251–262 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

