Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species
- PMID: 18753345
- PMCID: PMC2576583
- DOI: 10.1128/JCM.00927-08
Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species
Abstract
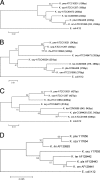
The 16S-23S rRNA gene internal transcribed spacer (ITS) regions of Klebsiella spp., including Klebsiella pneumoniae subsp. pneumoniae, Klebsiella pneumoniae subsp. ozaenae, Klebsiella pneumoniae subsp. rhinoscleromatis, Klebsiella oxytoca, Klebsiella planticola, Klebsiella terrigena, and Klebsiella ornithinolytica, were characterized, and the feasibility of using ITS sequences to discriminate Klebsiella species and subspecies was explored. A total of 336 ITS sequences from 21 representative strains and 11 clinical isolates of Klebsiella were sequenced and analyzed. Three distinct ITS types-ITS(none) (without tRNA genes), ITS(glu) [with a tRNA(Glu (UUC)) gene], and ITS(ile+ala) [with tRNA(Ile (GAU)) and tRNA(Ala (UGC)) genes]-were detected in all species except for K. pneumoniae subsp. rhinoscleromatis, which has only ITS(glu) and ITS(ile+ala). The presence of ITS(none) in Enterobacteriaceae had never been reported before. Both the length and the sequence of each ITS type are highly conserved within the species, with identity levels from 0.961 to 1.000 for ITS(none), from 0.967 to 1.000 for ITS(glu), and from 0.968 to 1.000 for ITS(ile+ala). Interspecies sequence identities range from 0.775 to 0.989 for ITS(none), from 0.798 to 0.997 for ITS(glu), and from 0.712 to 0.985 for ITS(ile+ala). Regions with significant interspecies variations but low intraspecies polymorphisms were identified; these may be targeted in the design of probes for the identification of Klebsiella to the species level. Phylogenetic analysis based on ITS regions reveals the relationships among Klebsiella species similarly to that based on 16S rRNA genes.
Figures




References
-
- Antón, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1998. Sequence diversity in the 16S-23S intergenic spacer region (ISR) of the rRNA operons in representatives of the Escherichia coli ECOR collection. J. Mol. Evol. 4762-72. - PubMed
-
- Barry, T., G. Colleran, M. Glennon, L. K. Dunican, and F. Gannon. 1991. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 151-56. - PubMed
-
- Biedenbach, D. J., G. J. Moet, and R. N. Jones. 2004. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002). Diagn. Microbiol. Infect. Dis. 5059-69. - PubMed
-
- Boye, K., and D. S. Hansen. 2003. Sequencing of 16S rDNA of Klebsiella: taxonomic relations within the genus and to other Enterobacteriaceae. Int. J. Med. Microbiol. 292495-503. - PubMed
-
- Boyer, S. L., V. R. Flechtner, and J. R. Johansen. 2001. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 181057-1069. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

