Factorial microarray analysis of zebrafish retinal development
- PMID: 18753621
- PMCID: PMC2529063
- DOI: 10.1073/pnas.0806038105
Factorial microarray analysis of zebrafish retinal development
Abstract
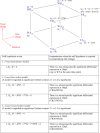
In a zebrafish recessive mutant young (yng), retinal cells are specified to distinct cell classes, but they fail to morphologically differentiate. A null mutation in a brahma-related gene 1 (brg1) is responsible for this phenotype. To identify retina-specific Brg1-regulated genes that control cellular differentiation, we conducted a factorial microarray analysis. Gene expression profiles were compared from wild-type and yng retinas and stage-matched whole embryos at 36 and 52 hours postfertilization (hpf). From our analysis, three categories of genes were identified: (i) Brg1-regulated retinal differentiation genes (731 probesets), (ii) retina-specific genes independent of Brg1 regulation (3,038 probesets), and (iii) Brg1-regulated genes outside the retina (107 probesets). Biological significance was confirmed by further analysis of components of the Cdk5 signaling pathway and Irx transcription factor family, representing genes identified in category 1. This study highlights the utility of factorial microarray analysis to efficiently identify relevant regulatory pathways influenced by both specific gene products and normal developmental events.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Leung YF, Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet. 2003;19:649–659. - PubMed
-
- Montgomery DC. Design and Analysis of Experiments. 6th Ed. Hoboken, NJ: Wiley; 2005.
-
- Wu C-F, Hamada M. Experiments: Planning, Analysis, and Parameter Design Optimization. New York: Wiley; 2000.
-
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. - PubMed
-
- Agathocleous M, Harris WA. Cell determination. In: Sernagor E, Eglen S, Harris B, Wong R, editors. Retinal Development. Cambridge: Cambridge Univ Press; 2006. pp. 75–98.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

