Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line
- PMID: 18753622
- PMCID: PMC2529074
- DOI: 10.1073/pnas.0806437105
Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line
Abstract
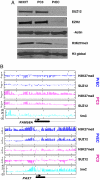
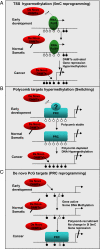
Epigenetic reprogramming is commonly observed in cancer, and is hypothesized to involve multiple mechanisms, including DNA methylation and Polycomb repressive complexes (PRCs). Here we devise a new experimental and analytical strategy using customized high-density tiling arrays to investigate coordinated patterns of gene expression, DNA methylation, and Polycomb marks which differentiate prostate cancer cells from their normal counterparts. Three major changes in the epigenomic landscape distinguish the two cell types. Developmentally significant genes containing CpG islands which are silenced by PRCs in the normal cells acquire DNA methylation silencing and lose their PRC marks (epigenetic switching). Because these genes are normally silent this switch does not cause de novo repression but might significantly reduce epigenetic plasticity. Two other groups of genes are silenced by either de novo DNA methylation without PRC occupancy (5mC reprogramming) or by de novo PRC occupancy without DNA methylation (PRC reprogramming). Our data suggest that the two silencing mechanisms act in parallel to reprogram the cancer epigenome and that DNA hypermethylation may replace Polycomb-based repression near key regulatory genes, possibly reducing their regulatory plasticity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Identification and functional relevance of de novo DNA methylation in cancerous B-cell populations.J Cell Biochem. 2010 Mar 1;109(4):818-27. doi: 10.1002/jcb.22461. J Cell Biochem. 2010. PMID: 20069569 Free PMC article.
-
Helicobacter pylori-infection-associated CpG island hypermethylation in the stomach and its possible association with polycomb repressive marks.Virchows Arch. 2008 May;452(5):515-24. doi: 10.1007/s00428-008-0596-7. Virchows Arch. 2008. PMID: 18335237
-
Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation.Cancer Res. 2009 Aug 1;69(15):6322-30. doi: 10.1158/0008-5472.CAN-09-0065. Epub 2009 Jul 14. Cancer Res. 2009. PMID: 19602592 Free PMC article.
-
Retinoids induce stem cell differentiation via epigenetic changes.Semin Cell Dev Biol. 2013 Dec;24(10-12):701-5. doi: 10.1016/j.semcdb.2013.08.002. Epub 2013 Aug 21. Semin Cell Dev Biol. 2013. PMID: 23973942 Free PMC article. Review.
-
DNA methylation patterns in lung carcinomas.Semin Cancer Biol. 2009 Jun;19(3):181-7. doi: 10.1016/j.semcancer.2009.02.008. Epub 2009 Feb 20. Semin Cancer Biol. 2009. PMID: 19429482 Free PMC article. Review.
Cited by
-
SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters.Mol Cancer Res. 2012 Oct;10(10):1332-42. doi: 10.1158/1541-7786.MCR-12-0351. Epub 2012 Sep 4. Mol Cancer Res. 2012. PMID: 22952045 Free PMC article.
-
Identification of DNA Methylation-Independent Epigenetic Events Underlying Clear Cell Renal Cell Carcinoma.Cancer Res. 2016 Apr 1;76(7):1954-64. doi: 10.1158/0008-5472.CAN-15-2622. Epub 2016 Jan 12. Cancer Res. 2016. PMID: 26759245 Free PMC article.
-
The role of 5-hydroxymethylcytosine in human cancer.Cell Tissue Res. 2014 Jun;356(3):631-41. doi: 10.1007/s00441-014-1896-7. Epub 2014 May 10. Cell Tissue Res. 2014. PMID: 24816989 Free PMC article. Review.
-
Genome-wide methylation analysis identifies a core set of hypermethylated genes in CIMP-H colorectal cancer.BMC Cancer. 2017 Mar 28;17(1):228. doi: 10.1186/s12885-017-3226-4. BMC Cancer. 2017. PMID: 28351398 Free PMC article.
-
Identification of the CIMP-like subtype and aberrant methylation of members of the chromosomal segregation and spindle assembly pathways in esophageal adenocarcinoma.Carcinogenesis. 2016 Apr;37(4):356-65. doi: 10.1093/carcin/bgw018. Epub 2016 Feb 10. Carcinogenesis. 2016. PMID: 26905591 Free PMC article.
References
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

