Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR
- PMID: 18755714
- PMCID: PMC2603282
- DOI: 10.1136/jcp.2008.058339
Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR
Abstract
Background: MicroRNAs have recently taken centre stage as short non-coding RNAs that regulate mRNA expression.
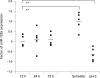
Aim/methods: To assess the feasibility of using microRNA techniques on routinely processed tissues, the accessibility of two representative microRNAs was examined by real-time quantitative PCR in 86 human formalin-fixed paraffin-embedded (FFPE) samples from liver, breast, bone marrow, lymphatic tissues and colon. Murine liver was used to analyse the influence of fixation time and different fixatives.
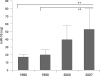
Results: High-quality microRNA was successfully extracted from routinely processed formalin-fixed tissues, resembling PCR amplification results from snap-frozen material analysed in parallel. While fixation time did not affect microRNA accessibility, non-buffered formalin or fixative supplements such as glutaraldehyde influenced PCR results. Storage of human tissues for up to 7 years did not cause a significant deterioration of microRNA. However, microRNA quality in human archival material following routine processing 10-20 years ago was decreased. Oxidation by ambient air during storage and fixation in non-buffered formalin is a possible reason for loss of microRNA quality.
Conclusion: The assessment of microRNAs in readily obtained formalin-fixed paraffin-embedded samples is a highly promising tool in molecular pathology when similarly treated samples are analysed. Therefore, microRNA analyses will gain wider acceptance as an adjunct to morphological tissue assessment in routine pathology and retrospective studies.
Conflict of interest statement
Figures


Similar articles
-
Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling.J Mol Diagn. 2008 Sep;10(5):415-23. doi: 10.2353/jmoldx.2008.080018. Epub 2008 Aug 7. J Mol Diagn. 2008. PMID: 18687792 Free PMC article.
-
Reliable microRNA profiling in routinely processed formalin-fixed paraffin-embedded breast cancer specimens using fluorescence labelled bead technology.BMC Biotechnol. 2008 Nov 27;8:90. doi: 10.1186/1472-6750-8-90. BMC Biotechnol. 2008. PMID: 19038028 Free PMC article.
-
Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues.J Mol Diagn. 2008 May;10(3):203-11. doi: 10.2353/jmoldx.2008.070153. Epub 2008 Apr 10. J Mol Diagn. 2008. PMID: 18403610 Free PMC article.
-
Excavation of a buried treasure--DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues.Histol Histopathol. 2011 Jun;26(6):797-810. doi: 10.14670/HH-26.797. Histol Histopathol. 2011. PMID: 21472693 Review.
-
Effects of fixative and fixation time on the extraction and polymerase chain reaction amplification of RNA from paraffin-embedded tissue. Comparison of two housekeeping gene mRNA controls.Diagn Mol Pathol. 1994 Sep;3(3):148-55. doi: 10.1097/00019606-199409000-00003. Diagn Mol Pathol. 1994. PMID: 7981889 Review.
Cited by
-
Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast.J Breast Cancer. 2011 Dec;14(4):269-75. doi: 10.4048/jbc.2011.14.4.269. Epub 2011 Dec 27. J Breast Cancer. 2011. PMID: 22323912 Free PMC article.
-
MicroRNA-29a-5p is a novel predictor for early recurrence of hepatitis B virus-related hepatocellular carcinoma after surgical resection.PLoS One. 2012;7(12):e52393. doi: 10.1371/journal.pone.0052393. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23285022 Free PMC article.
-
Expression of TRPM8 in human reactive lymphoid tissues and mature B-cell neoplasms.Oncol Lett. 2018 Nov;16(5):5930-5938. doi: 10.3892/ol.2018.9386. Epub 2018 Sep 3. Oncol Lett. 2018. PMID: 30344743 Free PMC article.
-
Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon.Virchows Arch. 2009 Jul;455(1):49-54. doi: 10.1007/s00428-009-0804-0. Epub 2009 Jun 23. Virchows Arch. 2009. PMID: 19547998
-
Changes of microRNAs-192, 196a and 203 correlate with Barrett's esophagus diagnosis and its progression compared to normal healthy individuals.Diagn Pathol. 2011 Nov 17;6:114. doi: 10.1186/1746-1596-6-114. Diagn Pathol. 2011. PMID: 22094011 Free PMC article.
References
-
- Rupp GM, Locker J. Purification and analysis of RNA from paraffin-embedded tissues. Biotechniques 1988;6:56–60 - PubMed
-
- Korbler T, Grskovic M, Dominis M, et al. A simple method for RNA isolation from formalin-fixed and paraffin-embedded lymphatic tissues. Exp Mol Pathol 2003;74:336–40 - PubMed
-
- Haque T, Faury D, Albrecht S, et al. Gene expression profiling from formalin-fixed paraffin-embedded tumors of pediatric glioblastoma. Clin Cancer Res 2007;13:6284–92 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
