Identification of pseudouridine methyltransferase in Escherichia coli
- PMID: 18755836
- PMCID: PMC2553739
- DOI: 10.1261/rna.1186608
Identification of pseudouridine methyltransferase in Escherichia coli
Abstract
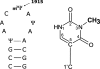
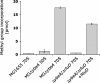
In ribosomal RNA, modified nucleosides are found in functionally important regions, but their function is obscure. Stem-loop 69 of Escherichia coli 23S rRNA contains three modified nucleosides: pseudouridines at positions 1911 and 1917, and N3 methyl-pseudouridine (m(3)Psi) at position 1915. The gene for pseudouridine methyltransferase was previously not known. We identified E. coli protein YbeA as the methyltransferase methylating Psi1915 in 23S rRNA. The E. coli ybeA gene deletion strain lacks the N3 methylation at position 1915 of 23S rRNA as revealed by primer extension and nucleoside analysis by HPLC. Methylation at position 1915 is restored in the ybeA deletion strain when recombinant YbeA protein is expressed from a plasmid. In addition, we show that purified YbeA protein is able to methylate pseudouridine in vitro using 70S ribosomes but not 50S subunits from the ybeA deletion strain as substrate. Pseudouridine is the preferred substrate as revealed by the inability of YbeA to methylate uridine at position 1915. This shows that YbeA is acting at the final stage during ribosome assembly, probably during translation initiation. Hereby, we propose to rename the YbeA protein to RlmH according to uniform nomenclature of RNA methyltransferases. RlmH belongs to the SPOUT superfamily of methyltransferases. RlmH was found to be well conserved in bacteria, and the gene is present in plant and in several archaeal genomes. RlmH is the first pseudouridine specific methyltransferase identified so far and is likely to be the only one existing in bacteria, as m(3)Psi1915 is the only methylated pseudouridine in bacteria described to date.
Figures



Similar articles
-
YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA.RNA. 2008 Oct;14(10):2234-44. doi: 10.1261/rna.1198108. Epub 2008 Aug 28. RNA. 2008. PMID: 18755835 Free PMC article.
-
Specificity and kinetics of 23S rRNA modification enzymes RlmH and RluD.RNA. 2010 Nov;16(11):2075-84. doi: 10.1261/rna.2234310. Epub 2010 Sep 3. RNA. 2010. PMID: 20817755 Free PMC article.
-
Different sensitivity of H69 modification enzymes RluD and RlmH to mutations in Escherichia coli 23S rRNA.Biochimie. 2012 May;94(5):1080-9. doi: 10.1016/j.biochi.2012.02.023. Biochimie. 2012. PMID: 22586702
-
Pseudouridines and pseudouridine synthases of the ribosome.Cold Spring Harb Symp Quant Biol. 2001;66:147-59. doi: 10.1101/sqb.2001.66.147. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762017 Review.
-
What do we know about ribosomal RNA methylation in Escherichia coli?Biochimie. 2015 Oct;117:110-8. doi: 10.1016/j.biochi.2014.11.019. Epub 2014 Dec 13. Biochimie. 2015. PMID: 25511423 Review.
Cited by
-
Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified.RNA. 2010 Oct;16(10):2023-32. doi: 10.1261/rna.2160010. Epub 2010 Aug 18. RNA. 2010. PMID: 20719918 Free PMC article.
-
A Family Divided: Distinct Structural and Mechanistic Features of the SpoU-TrmD (SPOUT) Methyltransferase Superfamily.Biochemistry. 2019 Feb 5;58(5):336-345. doi: 10.1021/acs.biochem.8b01047. Epub 2018 Dec 3. Biochemistry. 2019. PMID: 30457841 Free PMC article.
-
Pseudouridine and N1-methylpseudouridine as potent nucleotide analogues for RNA therapy and vaccine development.RSC Chem Biol. 2024 Mar 19;5(5):418-425. doi: 10.1039/d4cb00022f. eCollection 2024 May 8. RSC Chem Biol. 2024. PMID: 38725905 Free PMC article. Review.
-
Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae.Nucleic Acids Res. 2013 May 1;41(10):5428-43. doi: 10.1093/nar/gkt195. Epub 2013 Apr 4. Nucleic Acids Res. 2013. PMID: 23558746 Free PMC article.
-
METTL15 interacts with the assembly intermediate of murine mitochondrial small ribosomal subunit to form m4C840 12S rRNA residue.Nucleic Acids Res. 2020 Aug 20;48(14):8022-8034. doi: 10.1093/nar/gkaa522. Nucleic Acids Res. 2020. PMID: 32573735 Free PMC article.
References
-
- Ali, I.K., Lancaster, L., Feinberg, J., Joseph, S., Noller, H.F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. - PubMed
-
- Anantharaman, V., Koonin, E.V., Aravid, L. SPOUT: A class of methyltransferases that includes spoU and trmD RNA methylase superfamilies and novel superfamilies of predicted prokaryotic RNA methyltransferases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. - PubMed
-
- Andersen, N.M., Douthwaite, S. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
