Solution structure of the integral human membrane protein VDAC-1 in detergent micelles
- PMID: 18755977
- PMCID: PMC2579273
- DOI: 10.1126/science.1161302
Solution structure of the integral human membrane protein VDAC-1 in detergent micelles
Abstract
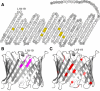
The voltage-dependent anion channel (VDAC) mediates trafficking of small molecules and ions across the eukaryotic outer mitochondrial membrane. VDAC also interacts with antiapoptotic proteins from the Bcl-2 family, and this interaction inhibits release of apoptogenic proteins from the mitochondrion. We present the nuclear magnetic resonance (NMR) solution structure of recombinant human VDAC-1 reconstituted in detergent micelles. It forms a 19-stranded beta barrel with the first and last strand parallel. The hydrophobic outside perimeter of the barrel is covered by detergent molecules in a beltlike fashion. In the presence of cholesterol, recombinant VDAC-1 can form voltage-gated channels in phospholipid bilayers similar to those of the native protein. NMR measurements revealed the binding sites of VDAC-1 for the Bcl-2 protein Bcl-x(L), for reduced beta-nicotinamide adenine dinucleotide, and for cholesterol. Bcl-x(L) interacts with the VDAC barrel laterally at strands 17 and 18.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

