Visualization, documentation, analysis, and communication of large-scale gene regulatory networks
- PMID: 18757046
- PMCID: PMC2762351
- DOI: 10.1016/j.bbagrm.2008.07.014
Visualization, documentation, analysis, and communication of large-scale gene regulatory networks
Abstract
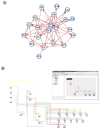
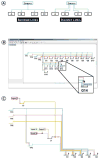
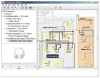
Genetic regulatory networks (GRNs) are complex, large-scale, and spatially and temporally distributed. These characteristics impose challenging demands on software tools for building GRN models, and so there is a need for custom tools. In this paper, we report on our ongoing development of BioTapestry, an open source, freely available computational tool designed specifically for building GRN models. We also outline our future development plans, and give some examples of current applications of BioTapestry.
Figures






Similar articles
-
BioTapestry: a tool to visualize the dynamic properties of gene regulatory networks.Methods Mol Biol. 2012;786:359-94. doi: 10.1007/978-1-61779-292-2_21. Methods Mol Biol. 2012. PMID: 21938637
-
Computational representation of developmental genetic regulatory networks.Dev Biol. 2005 Jul 1;283(1):1-16. doi: 10.1016/j.ydbio.2005.04.023. Dev Biol. 2005. PMID: 15907831 Review.
-
BioTapestry now provides a web application and improved drawing and layout tools.F1000Res. 2016 Jan 8;5:39. doi: 10.12688/f1000research.7620.1. eCollection 2016. F1000Res. 2016. PMID: 27134726 Free PMC article.
-
CABeRNET: a Cytoscape app for augmented Boolean models of gene regulatory NETworks.BMC Bioinformatics. 2016 Feb 4;17:64. doi: 10.1186/s12859-016-0914-z. BMC Bioinformatics. 2016. PMID: 26846964 Free PMC article.
-
Methods for the experimental and computational analysis of gene regulatory networks in sea urchins.Methods Cell Biol. 2019;151:89-113. doi: 10.1016/bs.mcb.2018.10.003. Epub 2018 Dec 11. Methods Cell Biol. 2019. PMID: 30948033 Review.
Cited by
-
A gene regulatory network controlling the embryonic specification of endoderm.Nature. 2011 May 29;474(7353):635-9. doi: 10.1038/nature10100. Nature. 2011. PMID: 21623371 Free PMC article.
-
Global analysis of photosynthesis transcriptional regulatory networks.PLoS Genet. 2014 Dec 11;10(12):e1004837. doi: 10.1371/journal.pgen.1004837. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25503406 Free PMC article.
-
Developmental single-cell transcriptomics in the Lytechinus variegatus sea urchin embryo.Development. 2021 Oct 1;148(19):dev198614. doi: 10.1242/dev.198614. Epub 2021 Sep 27. Development. 2021. PMID: 34463740 Free PMC article.
-
Developmental gene regulatory networks in sea urchins and what we can learn from them.F1000Res. 2016 Feb 22;5:F1000 Faculty Rev-203. doi: 10.12688/f1000research.7381.1. eCollection 2016. F1000Res. 2016. PMID: 26962438 Free PMC article. Review.
-
Feedback circuits are numerous in embryonic gene regulatory networks and offer a stabilizing influence on evolution of those networks.Evodevo. 2023 Jun 16;14(1):10. doi: 10.1186/s13227-023-00214-y. Evodevo. 2023. PMID: 37322563 Free PMC article.
References
-
- Longabaugh WJR, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. - PubMed
-
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The Systems Biology Markup Language (SBML): A Medium for Representation and Exchange of Biochemical Network Models. Bioinformatics. 2003;19(4):524–531. - PubMed
-
- Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, Longabaugh W, Vuthoori M, Whitehead K, Madar A, Suzuki L, Mori T, Chang D, DiRuggiero J, Johnson CH, Hood L, Baliga NS. A Predictive Model for Transcriptional Control of Physiology in a Free Living Cell. Cell. 2007;131:1354–1365. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

