Gly-Ala repeats induce position- and substrate-specific regulation of 26 S proteasome-dependent partial processing
- PMID: 18757367
- PMCID: PMC2662073
- DOI: 10.1074/jbc.M803290200
Gly-Ala repeats induce position- and substrate-specific regulation of 26 S proteasome-dependent partial processing
Abstract
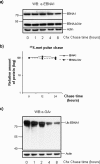
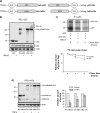
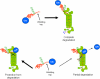
Partial degradation or regulated ubiquitin proteasome-dependent processing by the 26 S proteasome has been demonstrated, but the underlying molecular mechanisms and the prevalence of this phenomenon remain obscure. Here we show that the Gly-Ala repeat (GAr) sequence of EBNA1 affects processing of substrates via the ubiquitin-dependent degradation pathway in a substrate- and position-specific fashion. GAr-mediated increase in stability of proteins targeted for degradation via the 26 S proteasome was associated with a fraction of the substrates being partially processed and the release of the free GAr. The GAr did not cause a problem for the proteolytic activity of the proteasome, and its fusion to the N terminus of p53 resulted in an increase in the rate of degradation of the entire chimera. Interestingly the GAr had little effect on the stability of EBNA1 protein itself, and targeting EBNA1 for 26 S proteasome-dependent degradation led to its complete degradation. Taken together, our data suggest a model in which the GAr prevents degradation or promotes endoproteolytic processing of substrates targeted for the 26 S proteasome by interfering with the initiation step of substrate unfolding. These results will help to further understand the underlying mechanisms for partial proteasome-dependent degradation.
Figures




References
-
- Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373-428 - PubMed
-
- Elsasser, S., and Finley, D. (2005) Nat. Cell Biol. 7 742-749 - PubMed
-
- Rape, M., and Jentsch, S. (2002) Nat. Cell Biol. 4 E113-E116 - PubMed
-
- Hoppe, T., Matuschewski, K., Rape, M., Schlenker, S., Ulrich, H. D., and Jentsch, S. (2000) Cell 102 577-586 - PubMed
-
- Hoppe, T., Rape, M., and Jentsch, S. (2001) Curr. Opin. Cell Biol. 13 344-348 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

