Endoplasmic reticulum chaperone protein GRP-78 mediates endocytosis of dentin matrix protein 1
- PMID: 18757373
- PMCID: PMC2573078
- DOI: 10.1074/jbc.M800786200
Endoplasmic reticulum chaperone protein GRP-78 mediates endocytosis of dentin matrix protein 1
Abstract
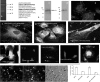
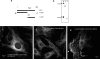
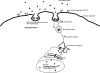
Dentin matrix protein 1 (DMP1), a phosphorylated protein present in the mineral phase of both vertebrates and invertebrates, is a key regulatory protein during biogenic formation of mineral deposits. Previously we showed that DMP1 is localized in the nuclear compartment of preosteoblasts and preodontoblasts. In the nucleus DMP1 might play an important role in the regulation of genes that control osteoblast or odontoblast differentiation. Here, we show that cellular uptake of DMP1 occurs through endocytosis. Interestingly, this process is initiated by DMP1 binding to the glucose-regulated protein-78 (GRP-78) localized on the plasma membrane of preodontoblast cells. Binding of DMP1 to GRP-78 receptor was determined to be specific and saturable with a binding dissociation constant K(D)=85 nm. We further depict a road map for the endocytosed DMP1 and demonstrate that the internalization is mediated primarily by caveolae and that the vesicles containing DMP1 are routed to the nucleus along microtubules. Immunohistochemical analysis and binding studies performed with biotin-labeled DMP1 confirm spatial co-localization of DMP1 and GRP-78 in the preodontoblasts of a developing mouse molar. Co-localization of DMP1 with GRP-78 was also observed in T4-4 preodontoblast cells, dental pulp stem cells, and primary preodontoblasts. By small interfering RNA techniques, we demonstrate that the receptor for DMP1 is GRP-78. Therefore, binding of DMP1 with GRP-78 receptor might be an important mechanism by which DMP1 is internalized and transported to the nucleus during bone and tooth development.
Figures







Similar articles
-
Stress chaperone GRP-78 functions in mineralized matrix formation.J Biol Chem. 2011 Mar 18;286(11):8729-39. doi: 10.1074/jbc.M110.179341. Epub 2011 Jan 14. J Biol Chem. 2011. PMID: 21239500 Free PMC article.
-
MMP2-cleavage of DMP1 generates a bioactive peptide promoting differentiation of dental pulp stem/progenitor cell.Eur Cell Mater. 2009 Nov 12;18:84-95. doi: 10.22203/ecm.v018a08. Eur Cell Mater. 2009. PMID: 19908197 Free PMC article.
-
Expression and distribution of grp-78/bip in mineralizing tissues and mesenchymal cells.Histochem Cell Biol. 2012 Jul;138(1):113-25. doi: 10.1007/s00418-012-0952-1. Epub 2012 Apr 17. Histochem Cell Biol. 2012. PMID: 22527697 Free PMC article.
-
Extracellular matrix proteins of dentine.Ciba Found Symp. 1997;205:107-15; discussion 115-7. doi: 10.1002/9780470515303.ch8. Ciba Found Symp. 1997. PMID: 9189620 Review.
-
The glucose-regulated proteins (GRP78 and GRP94): functions, gene regulation, and applications.Crit Rev Eukaryot Gene Expr. 1994;4(1):1-18. doi: 10.1615/critreveukargeneexpr.v4.i1.10. Crit Rev Eukaryot Gene Expr. 1994. PMID: 7987045 Review.
Cited by
-
Mitochondria chaperone GRP75 moonlighting as a cell cycle controller to derail endocytosis provides an opportunity for nanomicrosphere intracellular delivery.Oncotarget. 2017 Apr 19;8(35):58536-58552. doi: 10.18632/oncotarget.17234. eCollection 2017 Aug 29. Oncotarget. 2017. PMID: 28938577 Free PMC article.
-
Heat Shock Proteins in Tooth Development and Injury Repair.Int J Mol Sci. 2023 Apr 18;24(8):7455. doi: 10.3390/ijms24087455. Int J Mol Sci. 2023. PMID: 37108621 Free PMC article. Review.
-
Fine mapping of bone structure and strength QTLs in heterogeneous stock rat.Bone. 2015 Dec;81:417-426. doi: 10.1016/j.bone.2015.08.013. Epub 2015 Aug 19. Bone. 2015. PMID: 26297441 Free PMC article.
-
Copine-7 binds to the cell surface receptor, nucleolin, and regulates ciliogenesis and Dspp expression during odontoblast differentiation.Sci Rep. 2017 Sep 12;7(1):11283. doi: 10.1038/s41598-017-11641-y. Sci Rep. 2017. PMID: 28900213 Free PMC article.
-
Nucleus-targeted Dmp1 transgene fails to rescue dental defects in Dmp1 null mice.Int J Oral Sci. 2014 Sep;6(3):133-41. doi: 10.1038/ijos.2014.44. Epub 2014 Aug 8. Int J Oral Sci. 2014. PMID: 25105818 Free PMC article.
References
-
- Narayanan, K., Ramachandran, A., Hao, J., He, G., Park, K. W., Cho, M., and George, A. (2003) J. Biol. Chem. 278 17500–17508 - PubMed
-
- Narayanan, K., Gajjeraman, S., Ramachandran, A., Hao, J., and George, A. (2006) J. Biol. Chem. 281 19064–19071 - PubMed
-
- He, G., Dahl, T., Veis, A., and George, A. (2003) Nat. Mater., 2 552–558 - PubMed
-
- He, G., and George, A. (2004) J. Biol. Chem. 279 11649–11656 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

