EBV is necessary for proliferation of dually infected primary effusion lymphoma cells
- PMID: 18757410
- PMCID: PMC2587434
- DOI: 10.1158/0008-5472.CAN-08-0627
EBV is necessary for proliferation of dually infected primary effusion lymphoma cells
Abstract
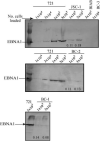
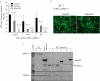
Epstein Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are found together in approximately 80% of primary effusion lymphomas (PEL), but their contribution to these cancers is unclear. We found that dominant-negative derivatives of EBNA1 inhibited EBV-positive PEL cells from forming colonies. Those rare PEL cells that proliferated after expression of the dominant-negative derivatives usually expressed these derivatives at low or undetectable levels and continued to maintain their EBV genomes. Those proliferating cells expressing higher levels of the derivatives expressed mutant derivatives that could not bind DNA. These findings indicate that EBV is required to sustain proliferation, as measured by colony formation of dually infected PEL cells. The dominant-negative derivatives of EBNA1 had no effect on the colony-forming ability of five EBV-negative, KSHV-negative hematopoietic cell lines. Surprisingly, they did inhibit the colony-forming ability of EBV-negative, KSHV-positive PEL cells. The small fraction of cells that continued to proliferate expressed only mutants of the EBNA1 derivatives that could no longer bind DNA. These findings indicate that the site-specific DNA-binding activity of EBNA1 or its derivatives when expressed efficiently in EBV-negative, KSHV-positive PEL cells inhibits their colony formation possibly through their binding to the KSHV genome.
Figures


References
-
- Jenner RG, Boshoff C. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim Biophys Acta. 2002;1602:1–22. - PubMed
-
- Walts AE, Shintaku IP, Said JW. Diagnosis of malignant lymphoma in effusions from patients with AIDS by gene rearrangement. Am J Clin Pathol. 1990;94:170–5. - PubMed
-
- Knowles DM, Inghirami G, Ubriaco A, Dalla-Favera R. Molecular genetic analysis of three AIDS-associated neoplasms of uncertain lineage demonstrates their B-cell derivation and the possible pathogenetic role of the Epstein-Barr virus. Blood. 1989;73:792–9. - PubMed
-
- Green I, Espiritu E, Ladanyi M, et al. Primary lymphomatous effusions in AIDS: a morphological, immunophenotypic, and molecular study. Mod Pathol. 1995;8:39–45. - PubMed
-
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

