EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's sarcoma
- PMID: 18757425
- PMCID: PMC4964910
- DOI: 10.1158/0008-5472.CAN-07-6145
EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's sarcoma
Abstract
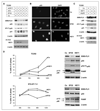
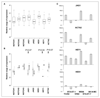
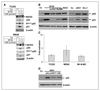
Although p53 is the most frequently mutated gene in cancer, half of human tumors retain wild-type p53, whereby it is unknown whether normal p53 function is compromised by other cancer-associated alterations. One example is Ewing's sarcoma family tumors (ESFT), where 90% express wild-type p53. ESFT are characterized by EWS-FLI1 oncogene fusions. Studying 6 ESFT cell lines, silencing of EWS-FLI1 in a wild-type p53 context resulted in increased p53 and p21(WAF1/CIP1) levels, causing cell cycle arrest. Using a candidate gene approach, HEY1 was linked to p53 induction. HEY1 was rarely expressed in 59 primary tumors, but consistently induced upon EWS-FLI1 knockdown in ESFT cell lines. The NOTCH signaling pathway targets HEY1, and we show NOTCH2 and NOTCH3 to be expressed in ESFT primary tumors and cell lines. Upon EWS-FLI1 silencing, NOTCH3 processing accompanied by nuclear translocation of the activated intracellular domain was observed in all but one p53-mutant cell line. In cell lines with the highest HEY1 induction, NOTCH3 activation was the consequence of JAG1 transcriptional induction. JAG1 modulation by specific siRNA, NOTCH-processing inhibition by either GSI or ectopic NUMB1, and siRNA-mediated HEY1 knockdown all inhibited p53 and p21(WAF1/CIP1) induction. Conversely, forced expression of JAG1, activated NOTCH3, or HEY1 induced p53 and p21(WAF1/CIP1). These results indicate that suppression of EWS-FLI1 reactivates NOTCH signaling in ESFT cells, resulting in p53-dependent cell cycle arrest. Our data link EWS-FLI1 to the NOTCH and p53 pathways and provide a plausible basis both for NOTCH tumor suppressor effects and oncogenesis of cancers that retain wild-type p53.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures


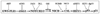
Similar articles
-
Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein.J Biol Chem. 2003 Apr 25;278(17):15105-15. doi: 10.1074/jbc.M211470200. Epub 2003 Jan 30. J Biol Chem. 2003. PMID: 12560328
-
A molecular function map of Ewing's sarcoma.PLoS One. 2009;4(4):e5415. doi: 10.1371/journal.pone.0005415. Epub 2009 Apr 30. PLoS One. 2009. PMID: 19404404 Free PMC article.
-
A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis.Dis Model Mech. 2012 Jan;5(1):95-106. doi: 10.1242/dmm.007401. Epub 2011 Oct 6. Dis Model Mech. 2012. PMID: 21979944 Free PMC article.
-
EWS-FLI1 in Ewing's sarcoma: real targets and collateral damage.Adv Exp Med Biol. 2006;587:41-52. doi: 10.1007/978-1-4020-5133-3_4. Adv Exp Med Biol. 2006. PMID: 17163154 Review.
-
Ewing's sarcoma oncoprotein EWS-FLI1: the perfect target without a therapeutic agent.Future Oncol. 2005 Aug;1(4):521-8. doi: 10.2217/14796694.1.4.521. Future Oncol. 2005. PMID: 16556028 Review.
Cited by
-
Target gene analysis by microarrays and chromatin immunoprecipitation identifies HEY proteins as highly redundant bHLH repressors.PLoS Genet. 2012;8(5):e1002728. doi: 10.1371/journal.pgen.1002728. Epub 2012 May 17. PLoS Genet. 2012. PMID: 22615585 Free PMC article.
-
Potential molecular targets for Ewing's sarcoma therapy.Indian J Med Paediatr Oncol. 2012 Oct;33(4):195-202. doi: 10.4103/0971-5851.107074. Indian J Med Paediatr Oncol. 2012. PMID: 23580819 Free PMC article.
-
Rethinking growth factors: the case of BMP9 during vessel maturation.Vasc Biol. 2022 Feb 7;4(1):R1-R14. doi: 10.1530/VB-21-0019. eCollection 2022 Feb 1. Vasc Biol. 2022. PMID: 35350597 Free PMC article. Review.
-
Advances in sarcoma genomics and new therapeutic targets.Nat Rev Cancer. 2011 Jul 14;11(8):541-57. doi: 10.1038/nrc3087. Nat Rev Cancer. 2011. PMID: 21753790 Free PMC article. Review.
-
Understanding the Biology of Bone Sarcoma from Early Initiating Events through Late Events in Metastasis and Disease Progression.Front Oncol. 2013 Sep 17;3:230. doi: 10.3389/fonc.2013.00230. Front Oncol. 2013. PMID: 24062983 Free PMC article. Review.
References
-
- Kovar H. Context matters: the hen or egg problem in Ewing’s sarcoma. Semin Cancer Biol. 2005;15:189–96. - PubMed
-
- Kovar H, Auinger A, Jug G, et al. Narrow spectrum of infrequent p53 mutations and absence of MDM2 amplification in Ewing tumours. Oncogene. 1993;8:2683–90. - PubMed
-
- De Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of P53 status in Ewing sarcoma. Cancer. 2000;89:783–92. - PubMed
-
- Kovar H, Jug G, Aryee DNT, et al. Among genes involved in the RB dependent cell cycle regulatory cascade, the p16 tumor suppressor gene is frequently lost in the Ewing family of tumors. Oncogene. 1997;15:2225–32. - PubMed
-
- Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. J Clin Oncol. 2005;23:548–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

