Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa Isolates from the ocean
- PMID: 18757570
- PMCID: PMC2570286
- DOI: 10.1128/AEM.02322-07
Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa Isolates from the ocean
Abstract
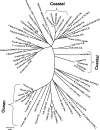
Recent isolation of Pseudomonas aeruginosa strains from the open ocean and subsequent pulsed-field gel electrophoresis analyses indicate that these strains have a unique genotype (N. H. Khan, Y. Ishii, N. Kimata-Kino, H. Esaki, T. Nishino, M. Nishimura, and K. Kogure, Microb. Ecol. 53:173-186, 2007). We hypothesized that ocean P. aeruginosa strains have a unique phylogenetic position relative to other strains. The objective of this study was to clarify the intraspecies phylogenetic relationship between marine strains and other strains from various geographical locations. Considering the advantages of using databases, multilocus sequence typing (MLST) was chosen for the typing and discrimination of ocean P. aeruginosa strains. Seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were analyzed, and the results were compared with data on the MLST website. These genes were also used for phylogenetic analysis of P. aeruginosa. Rooted and unrooted phylogenetic trees were generated for each gene locus and the concatenated gene fragments. MLST data showed that all the ocean strains were new. Trees constructed for individual and concatenated genes revealed that ocean P. aeruginosa strains have clusters distinct from those of other P. aeruginosa strains. These clusters roughly reflected the geographical locations of the isolates. These data support our previous findings that P. aeruginosa strains are present in the ocean. It can be concluded that the ocean P. aeruginosa strains have diverged from other isolates and form a distinct cluster based on MLST and phylogenetic analyses of seven housekeeping genes.
Figures



Similar articles
-
Multilocus sequence typing analysis of Pseudomonas aeruginosa isolated from pet Chinese stripe-necked turtles (Ocadia sinensis).Lab Anim Res. 2016 Dec;32(4):208-216. doi: 10.5625/lar.2016.32.4.208. Epub 2016 Dec 23. Lab Anim Res. 2016. PMID: 28053614 Free PMC article.
-
Multilocus sequencing typing of Pseudomonas aeruginosa isolates and analysis of potential pathogenicity of typical genotype strains from occupational oxyhelium saturation divers.Undersea Hyperb Med. 2014 Mar-Apr;41(2):135-41. Undersea Hyperb Med. 2014. PMID: 24851551
-
Genetic diversity of clinical Pseudomonas aeruginosa isolates in a public hospital in Spain.BMC Microbiol. 2013 Jun 18;13:138. doi: 10.1186/1471-2180-13-138. BMC Microbiol. 2013. PMID: 23773707 Free PMC article.
-
High-throughput single-nucleotide polymorphism-based typing of shared Pseudomonas aeruginosa strains in cystic fibrosis patients using the Sequenom iPLEX platform.J Med Microbiol. 2013 May;62(Pt 5):734-740. doi: 10.1099/jmm.0.055905-0. Epub 2013 Feb 14. J Med Microbiol. 2013. PMID: 23412772
-
Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: Practical overview.Microbiol Immunol. 2020 May;64(5):331-344. doi: 10.1111/1348-0421.12776. Epub 2020 Feb 11. Microbiol Immunol. 2020. PMID: 31965613 Review.
Cited by
-
Antimicrobial resistance and genomic rep-PCR fingerprints of Pseudomonas aeruginosa strains from animals on the background of the global population structure.BMC Vet Res. 2017 Feb 21;13(1):58. doi: 10.1186/s12917-017-0977-8. BMC Vet Res. 2017. PMID: 28222788 Free PMC article.
-
Determination of the diversity of Rhodopirellula isolates from European seas by multilocus sequence analysis.Appl Environ Microbiol. 2010 Feb;76(3):776-85. doi: 10.1128/AEM.01525-09. Epub 2009 Nov 30. Appl Environ Microbiol. 2010. PMID: 19948850 Free PMC article.
-
Differential habitat use and niche partitioning by Pseudomonas species in human homes.Microb Ecol. 2011 Oct;62(3):505-17. doi: 10.1007/s00248-011-9844-5. Epub 2011 Apr 19. Microb Ecol. 2011. PMID: 21503776
-
Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains.Front Microbiol. 2015 Aug 25;6:871. doi: 10.3389/fmicb.2015.00871. eCollection 2015. Front Microbiol. 2015. PMID: 26379646 Free PMC article.
-
Comparative genomic analysis of Shiga toxin-producing and non-Shiga toxin-producing Escherichia coli O157 isolated from outbreaks in Korea.Gut Pathog. 2017 Feb 6;9:7. doi: 10.1186/s13099-017-0156-2. eCollection 2017. Gut Pathog. 2017. PMID: 28191041 Free PMC article.
References
-
- Alonso, A., F. Rojo, and J. L. Martínez. 1999. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1:421-430. - PubMed
-
- Aoi, Y., H. Nakata, and H. Kida. 2000. Isolation of Pseudomonas aeruginosa from Ushubetsu River water in Hokkaido, Japan. Jpn. J. Vet. Res. 48:29-34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

