Novel Parachlamydia acanthamoebae quantification method based on coculture with amoebae
- PMID: 18757579
- PMCID: PMC2570282
- DOI: 10.1128/AEM.00841-08
Novel Parachlamydia acanthamoebae quantification method based on coculture with amoebae
Abstract
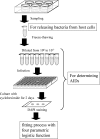
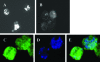
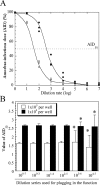
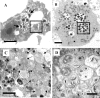
Parachlamydia acanthamoebae, belonging to the order Chlamydiales, is an obligately intracellular bacterium that infects free-living amoebae and is a potential human pathogen. However, no method exists to accurately quantify viable bacterial numbers. We present a novel quantification method for P. acanthamoebae based on coculture with amoebae. P. acanthamoebae was cultured either with Acanthamoeba spp. or with mammalian epithelial HEp-2 or Vero cells. The infection rate of P. acanthamoebae (amoeba-infectious dose [AID]) was determined by DAPI (4',6-diamidino-2-phenylindole) staining and was confirmed by fluorescent in situ hybridization. AIDs were plotted as logistic sigmoid dilution curves, and P. acanthamoebae numbers, defined as amoeba-infectious units (AIU), were calculated. During culture, amoeba numbers and viabilities did not change, and amoebae did not change from trophozoites to cysts. Eight amoeba strains showed similar levels of P. acanthamoebae growth, and bacterial numbers reached ca. 1,000-fold (10(9) AIU preculture) after 4 days. In contrast, no increase was observed for P. acanthamoebae in either mammalian cell line. However, aberrant structures in epithelial cells, implying possible persistent infection, were seen by transmission electron microscopy. Thus, our method could monitor numbers of P. acanthamoebae bacteria in host cells and may be useful for understanding chlamydiae present in the natural environment as human pathogens.
Figures


Similar articles
-
Host range of obligate intracellular bacterium Parachlamydia acanthamoebae.Microbiol Immunol. 2010 Nov;54(11):707-13. doi: 10.1111/j.1348-0421.2010.00265.x. Microbiol Immunol. 2010. PMID: 21155362
-
Impact of free-living amoebae on presence of Parachlamydia acanthamoebae in the hospital environment and its survival in vitro without requirement for amoebae.J Clin Microbiol. 2010 Sep;48(9):3360-5. doi: 10.1128/JCM.00366-10. Epub 2010 Jul 14. J Clin Microbiol. 2010. PMID: 20631104 Free PMC article.
-
Apoptosis Functions in Defense against Infection of Mammalian Cells with Environmental Chlamydiae.Infect Immun. 2020 May 20;88(6):e00851-19. doi: 10.1128/IAI.00851-19. Print 2020 May 20. Infect Immun. 2020. PMID: 32179584 Free PMC article.
-
Pathogenic potential of novel Chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria.Clin Microbiol Rev. 2006 Apr;19(2):283-97. doi: 10.1128/CMR.19.2.283-297.2006. Clin Microbiol Rev. 2006. PMID: 16614250 Free PMC article. Review.
-
Microorganisms resistant to free-living amoebae.Clin Microbiol Rev. 2004 Apr;17(2):413-33. doi: 10.1128/CMR.17.2.413-433.2004. Clin Microbiol Rev. 2004. PMID: 15084508 Free PMC article. Review.
Cited by
-
Differential Legionella spp. survival between intracellular and extracellular forms in thermal spring environments.Environ Sci Pollut Res Int. 2013 May;20(5):3098-106. doi: 10.1007/s11356-012-1159-7. Epub 2012 Oct 4. Environ Sci Pollut Res Int. 2013. PMID: 23054762
-
Acanthamoeba containing endosymbiotic chlamydia isolated from hospital environments and its potential role in inflammatory exacerbation.BMC Microbiol. 2016 Dec 15;16(1):292. doi: 10.1186/s12866-016-0906-1. BMC Microbiol. 2016. PMID: 27978822 Free PMC article.
-
Amoebal endosymbiont Parachlamydia acanthamoebae Bn9 can grow in immortal human epithelial HEp-2 cells at low temperature; an in vitro model system to study chlamydial evolution.PLoS One. 2015 Feb 2;10(2):e0116486. doi: 10.1371/journal.pone.0116486. eCollection 2015. PLoS One. 2015. PMID: 25643359 Free PMC article.
-
Draft Genome Sequence of High-Temperature-Adapted Protochlamydia sp. HS-T3, an Amoebal Endosymbiotic Bacterium Found in Acanthamoeba Isolated from a Hot Spring in Japan.Genome Announc. 2015 Feb 5;3(1):e01507-14. doi: 10.1128/genomeA.01507-14. Genome Announc. 2015. PMID: 25657277 Free PMC article.
-
Ciliates expel environmental Legionella-laden pellets to stockpile food.Appl Environ Microbiol. 2012 Aug;78(15):5247-57. doi: 10.1128/AEM.00421-12. Epub 2012 May 25. Appl Environ Microbiol. 2012. PMID: 22635991 Free PMC article.
References
-
- Al-Younes, H. M., T. Rudel, and T. F. Meyer. 1999. Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell. Microbiol. 1:237-247. - PubMed
-
- Armstrong, M. 2000. The pathogenesis of human Acanthamoeba infection. Infect. Dis. Rev. 2:65-73.
-
- Birtles, R. J., T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 29:925-926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

