The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning
- PMID: 18757750
- PMCID: PMC2533191
- DOI: 10.1073/pnas.0804276105
The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning
Abstract
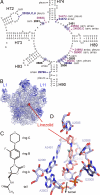
The oxazolidinones represent the first new class of antibiotics to enter into clinical usage within the past 30 years, but their binding site and mechanism of action has not been fully characterized. We have determined the crystal structure of the oxazolidinone linezolid bound to the Deinococcus radiodurans 50S ribosomal subunit. Linezolid binds in the A site pocket at the peptidyltransferase center of the ribosome overlapping the aminoacyl moiety of an A-site bound tRNA as well as many clinically important antibiotics. Binding of linezolid stabilizes a distinct conformation of the universally conserved 23S rRNA nucleotide U2585 that would be nonproductive for peptide bond formation. In conjunction with available biochemical data, we present a model whereby oxazolidinones impart their inhibitory effect by perturbing the correct positioning of tRNAs on the ribosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Spahn CMT, Prescott CD. Throwing a spanner in the works: Antibiotics and the translational apparatus. J Mol Med. 1996;74:423–439. - PubMed
-
- Wilson DN. In: Protein Synthesis and Ribosome Structure. Nierhaus K, Wilson DN, editors. Weinheim: Wiley–VCH; 2004. pp. 449–527.
-
- Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. - PubMed
-
- Tu D, Blaha G, Moore P, Steitz T. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

