A misexpression screen to identify regulators of Drosophila larval hemocyte development
- PMID: 18757933
- PMCID: PMC2535679
- DOI: 10.1534/genetics.108.089094
A misexpression screen to identify regulators of Drosophila larval hemocyte development
Abstract
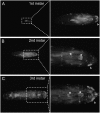
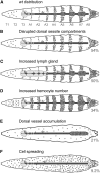
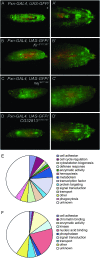
In Drosophila, defense against foreign pathogens is mediated by an effective innate immune system, the cellular arm of which is composed of circulating hemocytes that engulf bacteria and encapsulate larger foreign particles. Three hemocyte types occur: plasmatocytes, crystal cells, and lamellocytes. The most abundant larval hemocyte type is the plasmatocyte, which is responsible for phagocytosis and is present either in circulation or in adherent sessile domains under the larval cuticle. The mechanisms controlling differentiation of plasmatocytes and their migration toward these sessile compartments are unclear. To address these questions we have conducted a misexpression screen using the plasmatocyte-expressed GAL4 driver Peroxidasin-GAL4 (Pxn-GAL4) and existing enhancer-promoter (EP) and EP yellow (EY) transposon libraries to systematically misexpress approximately 20% of Drosophila genes in larval hemocytes. The Pxn-GAL4 strain also contains a UAS-GFP reporter enabling hemocyte phenotypes to be visualized in the semitransparent larvae. Among 3412 insertions screened we uncovered 101 candidate hemocyte regulators. Some of these are known to control hemocyte development, but the majority either have no characterized function or are proteins of known function not previously implicated in hemocyte development. We have further analyzed three candidate genes for changes in hemocyte morphology, cell-cell adhesion properties, phagocytosis activity, and melanotic tumor formation.
Figures



Similar articles
-
Lineage tracing of lamellocytes demonstrates Drosophila macrophage plasticity.PLoS One. 2010 Nov 19;5(11):e14051. doi: 10.1371/journal.pone.0014051. PLoS One. 2010. PMID: 21124962 Free PMC article.
-
Genetic Screen in Drosophila Larvae Links ird1 Function to Toll Signaling in the Fat Body and Hemocyte Motility.PLoS One. 2016 Jul 28;11(7):e0159473. doi: 10.1371/journal.pone.0159473. eCollection 2016. PLoS One. 2016. PMID: 27467079 Free PMC article.
-
An in vivo RNA interference screen identifies gene networks controlling Drosophila melanogaster blood cell homeostasis.BMC Dev Biol. 2010 Jun 11;10:65. doi: 10.1186/1471-213X-10-65. BMC Dev Biol. 2010. PMID: 20540764 Free PMC article.
-
Drosophila immune cell migration and adhesion during embryonic development and larval immune responses.Curr Opin Cell Biol. 2015 Oct;36:71-9. doi: 10.1016/j.ceb.2015.07.003. Epub 2015 Jul 24. Curr Opin Cell Biol. 2015. PMID: 26210104 Review.
-
Ontogeny of the Drosophila larval hematopoietic organ, hemocyte homeostasis and the dedicated cellular immune response to parasitism.Int J Dev Biol. 2010;54(6-7):1117-25. doi: 10.1387/ijdb.093053jk. Int J Dev Biol. 2010. PMID: 20711989 Review.
Cited by
-
A mosquito juvenile hormone binding protein (mJHBP) regulates the activation of innate immune defenses and hemocyte development.PLoS Pathog. 2020 Jan 21;16(1):e1008288. doi: 10.1371/journal.ppat.1008288. eCollection 2020 Jan. PLoS Pathog. 2020. PMID: 31961911 Free PMC article.
-
The raspberry Gene Is Involved in the Regulation of the Cellular Immune Response in Drosophila melanogaster.PLoS One. 2016 Mar 4;11(3):e0150910. doi: 10.1371/journal.pone.0150910. eCollection 2016. PLoS One. 2016. PMID: 26942456 Free PMC article.
-
Drosophila Genotype Influences Commensal Bacterial Levels.PLoS One. 2017 Jan 17;12(1):e0170332. doi: 10.1371/journal.pone.0170332. eCollection 2017. PLoS One. 2017. PMID: 28095502 Free PMC article.
-
Macrophage-like Blood Cells Are Involved in Inter-Tissue Communication to Activate JAK/STAT Signaling, Inducing Antitumor Turandot Proteins in Drosophila Fat Body via the TNF-JNK Pathway.Int J Mol Sci. 2024 Dec 6;25(23):13110. doi: 10.3390/ijms252313110. Int J Mol Sci. 2024. PMID: 39684820 Free PMC article.
-
Abrupt-mediated control of ninjurins regulates Drosophila sessile haemocyte compartments.Development. 2024 Dec 1;151(23):dev202977. doi: 10.1242/dev.202977. Epub 2024 Dec 9. Development. 2024. PMID: 39545919 Free PMC article.
References
-
- Abrams, J. M., K. White, L. I. Fessler and H. Steller, 1993. Programmed cell death during Drosophila embryogenesis. Development 117 29–43. - PubMed
-
- Alfonso, T. B., and B. W. Jones, 2002. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev. Biol. 248 369–383. - PubMed
-
- Baker, N. E., and A. E. Zitron, 1995. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech. Dev. 49 173–189. - PubMed
-
- Barrett, K., M. Leptin and J. Settleman, 1997. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell 91 905–915. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

