A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery
- PMID: 18760393
- PMCID: PMC2556439
- DOI: 10.1016/j.ajhg.2008.08.001
A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery
Abstract
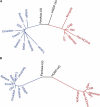
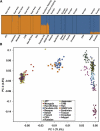
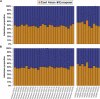
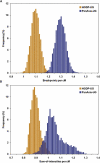
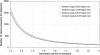
Following up on our previous study, we conducted a genome-wide analysis of admixture for two Uyghur population samples (HGDP-UG and PanAsia-UG), collected from the northern and southern regions of Xinjiang in China, respectively. Both HGDP-UG and PanAsia-UG showed a substantial admixture of East-Asian (EAS) and European (EUR) ancestries, with an empirical estimation of ancestry contribution of 53:47 (EAS:EUR) and 48:52 for HGDP-UG and PanAsia-UG, respectively. The effective admixture time under a model with a single pulse of admixture was estimated as 110 generations and 129 generations, or admixture events occurred about 2200 and 2580 years ago for HGDP-UG and PanAsia-UG, respectively, assuming an average of 20 yr per generation. Despite Uyghurs' earlier history compared to other admixture populations, admixture mapping, holds promise for this population, because of its large size and its mixture of ancestry from different continents. We screened multiple databases and identified a genome-wide single-nucleotide polymorphism panel that can distinguish EAS and EUR ancestry of chromosomal segments in Uyghurs. The panel contains 8150 ancestry-informative markers (AIMs) showing large frequency differences between EAS and EUR populations (F(ST) > 0.25, mean F(ST) = 0.43) but small frequency differences (7999 AIMs validated) within both populations (F(ST) < 0.05, mean F(ST) < 0.01). We evaluated the effectiveness of this admixture map for localizing disease genes in two Uyghur populations. To our knowledge, our map constitutes the first practical resource for admixture mapping in Uyghurs, and it will enable studies of diseases showing differences in genetic risk between EUR and EAS populations.
Figures
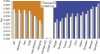


Comment in
-
Genetic landscape of Eurasia and "admixture" in Uyghurs.Am J Hum Genet. 2009 Dec;85(6):934-7; author reply 937-9. doi: 10.1016/j.ajhg.2009.10.024. Am J Hum Genet. 2009. PMID: 20004770 Free PMC article. No abstract available.
References
-
- Ai Q., Xiao H., Zhao J., Xu Y., Shi F. A survey on physical characteristics of Uigur Nationality. ACTA Anthropologica Sinica. 1993;12:357–365.
-
- Yao Y.G., Kong Q.P., Wang C.Y., Zhu C.L., Zhang Y.P. Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china. Mol. Biol. Evol. 2004;21:2265–2280. - PubMed
-
- Du R. Human Population Genetics Studies in China. Bulletin of Biology. 1997;32:9–12.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

