Non-cell-autonomous inhibition of photoreceptor development by Dip3
- PMID: 18761008
- PMCID: PMC2614221
- DOI: 10.1016/j.ydbio.2008.08.004
Non-cell-autonomous inhibition of photoreceptor development by Dip3
Abstract
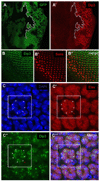
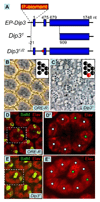
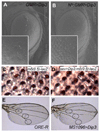
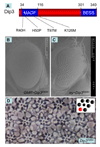
We show here that the Drosophila MADF/BESS domain transcription factor Dip3, which is expressed in differentiating photoreceptors, regulates neuronal differentiation in the compound eye. Loss of Dip3 activity in photoreceptors leads to an extra photoreceptor in many ommatidia, while ectopic expression of Dip3 in non-neuronal cells results in photoreceptor loss. These findings are consistent with the idea that Dip3 is required non-cell autonomously to block extra photoreceptor formation. Dip3 may mediate the spatially restricted potentiation of Notch (N) signaling since the Dip3 misexpression phenotype is suppressed by reducing N signaling and misexpression of Dip3 leads to ectopic activity of a N-responsive enhancer. Analysis of mosaic ommatidia suggests that no specific photoreceptor must be mutant to generate the mutant phenotype. Remarkably, however, mosaic pupal ommatidia with three or fewer Dip3(+) photoreceptors always differentiate an extra photoreceptor, while those with four or more Dip3(+) photoreceptors never differentiate an extra photoreceptor. These findings are consistent with the notion that Dip3 in photoreceptors activates a heretofore unsuspected diffusible ligand that may work in conjunction with the N pathway to prevent a subpopulation of undifferentiated cells from choosing a neuronal fate.
Figures

Similar articles
-
Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye.Dev Biol. 2005 Sep 1;285(1):38-48. doi: 10.1016/j.ydbio.2005.05.038. Dev Biol. 2005. PMID: 16039641
-
Control of photoreceptor cell morphology, planar polarity and epithelial integrity during Drosophila eye development.Development. 2002 May;129(9):2247-58. doi: 10.1242/dev.129.9.2247. Development. 2002. PMID: 11959832
-
Dorsal interacting protein 3 potentiates activation by Drosophila Rel homology domain proteins.Dev Comp Immunol. 2008;32(11):1290-300. doi: 10.1016/j.dci.2008.04.006. Epub 2008 May 16. Dev Comp Immunol. 2008. PMID: 18538389 Free PMC article.
-
Signal integration during development: insights from the Drosophila eye.Dev Dyn. 2004 Jan;229(1):162-75. doi: 10.1002/dvdy.10449. Dev Dyn. 2004. PMID: 14699588 Review.
-
Eye development: Notch lends a handedness.Curr Biol. 1999 May 20;9(10):R356-60. doi: 10.1016/s0960-9822(99)80226-7. Curr Biol. 1999. PMID: 10339422 Review.
Cited by
-
Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge.Genetics. 2014 Feb;196(2):481-96. doi: 10.1534/genetics.113.160531. Epub 2013 Dec 13. Genetics. 2014. PMID: 24336749 Free PMC article.
-
Stonewall and Brickwall: Two Partially Redundant Determinants Required for the Maintenance of Female Germline in Drosophila.G3 (Bethesda). 2018 May 31;8(6):2027-2041. doi: 10.1534/g3.118.200192. G3 (Bethesda). 2018. PMID: 29669801 Free PMC article.
References
-
- Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. - PubMed
-
- Baker NE, Zitron AE. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech Dev. 1995;49:173–189. - PubMed
-
- Banerjee U, Renfranz PJ, Pollock JA, Benzer S. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell. 1987;49:281–291. - PubMed
-
- Basler K, Hafen E. Specification of cell fate in the developing eye of Drosophila. Bioessays. 1991;13:621–631. - PubMed
-
- Blochlinger K, Jan LY, Jan YN. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

