A conserved anti-repressor controls horizontal gene transfer by proteolysis
- PMID: 18761623
- PMCID: PMC2581648
- DOI: 10.1111/j.1365-2958.2008.06414.x
A conserved anti-repressor controls horizontal gene transfer by proteolysis
Abstract
The mobile genetic element ICEBs1 is an integrative and conjugative element (a conjugative transposon) found in the Bacillus subtilis chromosome. The SOS response and the RapI-PhrI sensory system activate ICEBs1 gene expression, excision and transfer by inactivating the ICEBs1 repressor protein ImmR. Although ImmR is similar to many characterized phage repressors, we found that, unlike these repressors, inactivation of ImmR requires an ICEBs1-encoded anti-repressor ImmA (YdcM). ImmA was needed for the degradation of ImmR in B. subtilis. Coexpression of ImmA and ImmR in Escherichia coli or co-incubation of purified ImmA and ImmR resulted in site-specific cleavage of ImmR. Homologues of immR and immA are found in many mobile genetic elements. We found that the ImmA homologue encoded by B. subtilis phage phi105 is required for inactivation of the phi105 repressor (an ImmR homologue). ImmA-dependent proteolysis of ImmR repressors may be a conserved mechanism for regulating horizontal gene transfer.
Figures

 ). IPTG was present throughout growth of JMA840. D. immA+ (JMA836, □); ΔimmA (JMA838, τ); ΔimmA Pspank-immA (JMA842, ∇).
). IPTG was present throughout growth of JMA840. D. immA+ (JMA836, □); ΔimmA (JMA838, τ); ΔimmA Pspank-immA (JMA842, ∇).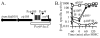
 , ø1050/R+A+). Cells were grown in minimal medium containing IPTG and were treated with mitomycin C at OD600 ∼ 0.5. Samples were collected at the times indicated and β-galactosidase specific activity was determined.
, ø1050/R+A+). Cells were grown in minimal medium containing IPTG and were treated with mitomycin C at OD600 ∼ 0.5. Samples were collected at the times indicated and β-galactosidase specific activity was determined.

Comment in
-
Back to the future: the new ICE age.Mol Microbiol. 2008 Nov;70(3):554-6. doi: 10.1111/j.1365-2958.2008.06415.x. Epub 2008 Aug 25. Mol Microbiol. 2008. PMID: 18761693
References
-
- Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, O'Brien TC, Shah A, Tierney JT, Tomm LL, O'Gara TM, Goranov AI, Grossman AD, Lovett CM. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187:7655–7666. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

