Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB
- PMID: 18761695
- PMCID: PMC2579741
- DOI: 10.1111/j.1365-2958.2008.06404.x
Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB
Abstract
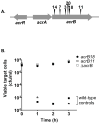
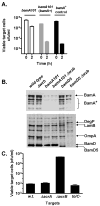
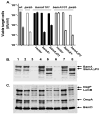
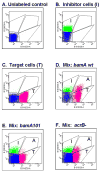
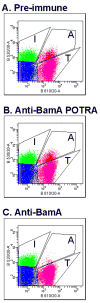
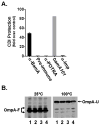
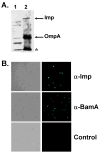
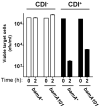
Contact-dependent growth inhibition (CDI) is a phenomenon by which bacterial cell growth is regulated by direct cell-to-cell contact via the CdiA/CdiB two-partner secretion system. Characterization of mutants resistant to CDI allowed us to identify BamA (YaeT) as the outer membrane receptor for CDI and AcrB as a potential downstream target. Notably, both BamA and AcrB are part of distinct multi-component machines. The Bam machine assembles outer membrane beta-barrel proteins into the outer membrane and the Acr machine exports small molecules into the extracellular milieu. We discovered that a mutation that reduces expression of BamA decreased binding of CDI+ inhibitor cells, measured by flow cytometry with fluorescently labelled bacteria. In addition, alpha-BamA antibodies, which recognized extracellular epitopes of BamA based on immunofluorescence, specifically blocked inhibitor-target cells binding and CDI. A second class of CDI-resistant mutants identified carried null mutations in the acrB gene. AcrB is an inner membrane component of a multidrug efflux pump that normally forms a cell envelope-spanning complex with the membrane fusion protein AcrA and the outer membrane protein TolC. Strikingly, the requirement for the BamA and AcrB proteins in CDI is independent of their multi-component machines, and thus their role in the CDI pathway may reflect novel, import-related functions.
Figures


References
-
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. - PubMed
-
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. - PubMed
-
- Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

