The emerging genetic architecture of type 2 diabetes
- PMID: 18762020
- PMCID: PMC4267677
- DOI: 10.1016/j.cmet.2008.08.006
The emerging genetic architecture of type 2 diabetes
Abstract
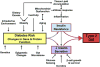
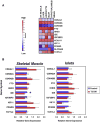
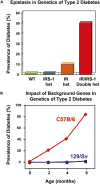
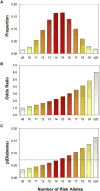
Type 2 diabetes is a genetically heterogeneous disease, with several relatively rare monogenic forms and a number of more common forms resulting from a complex interaction of genetic and environmental factors. Previous studies using a candidate gene approach, family linkage studies, and gene expression profiling uncovered a number of type 2 genes, but the genetic basis of common type 2 diabetes remained unknown. Recently, a new window has opened on defining potential type 2 diabetes genes through genome-wide SNP association studies of very large populations of individuals with diabetes. This review explores the pathway leading to discovery of these genetic effects, the impact of these genetic loci on diabetes risk, the potential mechanisms of action of the genes to alter glucose homeostasis, and the limitations of these studies in defining the role of genetics in this important disease.
Figures
References
-
- Almind K, Kulkarni RN, Lannon SM, Kahn CR. Identification of Interactive Loci Linked to Insulin and Leptin in Mice With Genetic Insulin Resistance. Diabetes. 2003;52:1535–1543. - PubMed
-
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al. The common PPARgamma Pro12Ala is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. - PubMed
-
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

