The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression
- PMID: 18762579
- PMCID: PMC2528585
- DOI: 10.1083/jcb.200803098
The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression
Abstract
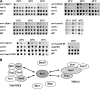
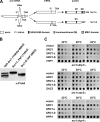
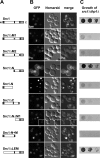
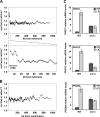
Inner nuclear membrane proteins containing a LEM (LAP2, emerin, and MAN1) domain participate in different processes, including chromatin organization, gene expression, and nuclear envelope biogenesis. In this study, we identify a robust genetic interaction between transcription export (TREX) factors and yeast Src1, an integral inner nuclear membrane protein that is homologous to vertebrate LEM2. DNA macroarray analysis revealed that the expression of the phosphate-regulated genes PHO11, PHO12, and PHO84 is up-regulated in src1Delta cells. Notably, these PHO genes are located in subtelomeric regions of chromatin and exhibit a perinuclear location in vivo. Src1 spans the nuclear membrane twice and exposes its N and C domains with putative DNA-binding motifs to the nucleoplasm. Genome-wide chromatin immunoprecipitation-on-chip analyses indicated that Src1 is highly enriched at telomeres and subtelomeric regions of the yeast chromosomes. Our data show that the inner nuclear membrane protein Src1 functions at the interface between subtelomeric gene expression and TREX-dependent messenger RNA export through the nuclear pore complexes.
Figures





References
-
- Akhtar, A., and S.M. Gasser. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517. - PubMed
-
- Alberola, T.M., J. García-Martínez, O. Antúnez, L. Viladevall, A. Barcelo, J. Ariño, and J.E. Pérez-Ortín. 2004. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 7:199–206. - PubMed
-
- Brachner, A., S. Reipert, R. Foisner, and J. Gotzmann. 2005. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 118:5797–5810. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

